Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung
- PMID: 26330513
- PMCID: PMC4556809
- DOI: 10.1128/mBio.00981-15
Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung
Abstract
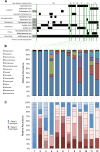
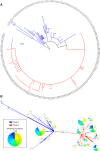
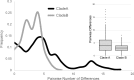
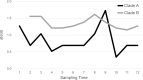
Pulmonary infections caused by Pseudomonas aeruginosa are a recalcitrant problem in cystic fibrosis (CF) patients. While the clinical implications and long-term evolutionary patterns of these infections are well studied, we know little about the short-term population dynamics that enable this pathogen to persist despite aggressive antimicrobial therapy. Here, we describe a short-term population genomic analysis of 233 P. aeruginosa isolates collected from 12 sputum specimens obtained over a 1-year period from a single patient. Whole-genome sequencing and antimicrobial susceptibility profiling identified the expansion of two clonal lineages. The first lineage originated from the coalescence of the entire sample less than 3 years before the end of the study and gave rise to a high-diversity ancestral population. The second expansion occurred 2 years later and gave rise to a derived population with a strong signal of positive selection. These events show characteristics consistent with recurrent selective sweeps. While we cannot identify the specific mutations responsible for the origins of the clonal lineages, we find that the majority of mutations occur in loci previously associated with virulence and resistance. Additionally, approximately one-third of all mutations occur in loci that are mutated multiple times, highlighting the importance of parallel pathoadaptation. One such locus is the gene encoding penicillin-binding protein 3, which received three independent mutations. Our functional analysis of these alleles shows that they provide differential fitness benefits dependent on the antibiotic under selection. These data reveal that bacterial populations can undergo extensive and dramatic changes that are not revealed by lower-resolution analyses.
Importance: Pseudomonas aeruginosa is a bacterial opportunistic pathogen responsible for significant morbidity and mortality in cystic fibrosis (CF) patients. Once it has colonized the lung in CF, it is highly resilient and rarely eradicated. This study presents a deep sampling examination of the fine-scale evolutionary dynamics of P. aeruginosa in the lungs of a chronically infected CF patient. We show that diversity of P. aeruginosa is driven by recurrent clonal emergence and expansion within this patient and identify potential adaptive variants associated with these events. This high-resolution sequencing strategy thus reveals important intraspecies dynamics that explain a clinically important phenomenon not evident at a lower-resolution analysis of community structure.
Copyright © 2015 Diaz Caballero et al.
Figures

References
-
- Tunney MM, Klem ER, Fodor AA, Gilpin DF, Moriarty TF, McGrath SJ, Muhlebach MS, Boucher RC, Cardwell C, Doering G, Elborn JS, Wolfgang MC. 2011. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 66:579–584. doi:10.1136/thx.2010.137281. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

