The Social Amoeba Dictyostelium discoideum Is Highly Resistant to Polyglutamine Aggregation
- PMID: 26330554
- PMCID: PMC4646202
- DOI: 10.1074/jbc.M115.676247
The Social Amoeba Dictyostelium discoideum Is Highly Resistant to Polyglutamine Aggregation
Abstract
The expression, misfolding, and aggregation of long repetitive amino acid tracts are a major contributing factor in a number of neurodegenerative diseases, including C9ORF72 amyotrophic lateral sclerosis/frontotemporal dementia, fragile X tremor ataxia syndrome, myotonic dystrophy type 1, spinocerebellar ataxia type 8, and the nine polyglutamine diseases. Protein aggregation is a hallmark of each of these diseases. In model organisms, including yeast, worms, flies, mice, rats, and human cells, expression of proteins with the long repetitive amino acid tracts associated with these diseases recapitulates the protein aggregation that occurs in human disease. Here we show that the model organism Dictyostelium discoideum has evolved to normally encode long polyglutamine tracts and express these proteins in a soluble form. We also show that Dictyostelium has the capacity to suppress aggregation of a polyglutamine-expanded Huntingtin construct that aggregates in other model organisms tested. Together, these data identify Dictyostelium as a novel model organism with the capacity to suppress aggregation of proteins with long polyglutamine tracts.
Keywords: Dictyostelium; neurodegenerative disease; polyglutamine; protein aggregation; protein folding.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


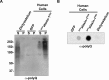
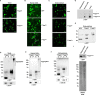
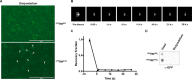
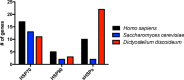
Similar articles
-
SRCP1 Conveys Resistance to Polyglutamine Aggregation.Mol Cell. 2018 Jul 19;71(2):216-228.e7. doi: 10.1016/j.molcel.2018.07.008. Mol Cell. 2018. PMID: 30029002 Free PMC article.
-
Mechanistic Insight into the Suppression of Polyglutamine Aggregation by SRCP1.ACS Chem Biol. 2023 Mar 17;18(3):549-560. doi: 10.1021/acschembio.2c00893. Epub 2023 Feb 15. ACS Chem Biol. 2023. PMID: 36791332 Free PMC article.
-
Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum.PLoS Genet. 2011 Apr;7(4):e1002052. doi: 10.1371/journal.pgen.1002052. Epub 2011 Apr 28. PLoS Genet. 2011. PMID: 21552328 Free PMC article.
-
Insights on Microsatellite Characteristics, Evolution, and Function From the Social Amoeba Dictyostelium discoideum.Front Neurosci. 2022 Jun 13;16:886837. doi: 10.3389/fnins.2022.886837. eCollection 2022. Front Neurosci. 2022. PMID: 35769695 Free PMC article. Review.
-
Drosophila melanogaster As a Model Organism to Study RNA Toxicity of Repeat Expansion-Associated Neurodegenerative and Neuromuscular Diseases.Front Cell Neurosci. 2017 Mar 21;11:70. doi: 10.3389/fncel.2017.00070. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28377694 Free PMC article. Review.
Cited by
-
Eukaryotic life without tQCUG: the role of Elongator-dependent tRNA modifications in Dictyostelium discoideum.Nucleic Acids Res. 2020 Aug 20;48(14):7899-7913. doi: 10.1093/nar/gkaa560. Nucleic Acids Res. 2020. PMID: 32609816 Free PMC article.
-
SRCP1 Conveys Resistance to Polyglutamine Aggregation.Mol Cell. 2018 Jul 19;71(2):216-228.e7. doi: 10.1016/j.molcel.2018.07.008. Mol Cell. 2018. PMID: 30029002 Free PMC article.
-
Viral vector gene delivery of the novel chaperone protein SRCP1 to modify insoluble protein in in vitro and in vivo models of ALS.Gene Ther. 2023 Jun;30(6):528-533. doi: 10.1038/s41434-021-00276-4. Epub 2021 Jul 8. Gene Ther. 2023. PMID: 34239068 Free PMC article.
-
Identification of Low-Complexity Domains by Compositional Signatures Reveals Class-Specific Frequencies and Functions Across the Domains of Life.PLoS Comput Biol. 2024 May 15;20(5):e1011372. doi: 10.1371/journal.pcbi.1011372. eCollection 2024 May. PLoS Comput Biol. 2024. PMID: 38748749 Free PMC article.
-
Using the social amoeba Dictyostelium to study the functions of proteins linked to neuronal ceroid lipofuscinosis.J Biomed Sci. 2016 Nov 24;23(1):83. doi: 10.1186/s12929-016-0301-0. J Biomed Sci. 2016. PMID: 27881166 Free PMC article. Review.
References
-
- Duennwald M. L. (2013) Yeast as a platform to explore polyglutamine toxicity and aggregation. Methods Mol. Biol. 1017, 153–161 - PubMed
-
- Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. (1998) Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95, 41–53 - PubMed
-
- Paulson H. L., Perez M. K., Trottier Y., Trojanowski J. Q., Subramony S. H., Das S. S., Vig P., Mandel J. L., Fischbeck K. H., Pittman R. N. (1997) Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

