Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27
- PMID: 26330557
- PMCID: PMC4646281
- DOI: 10.1074/jbc.M115.680942
Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27
Abstract
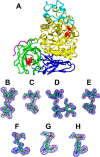
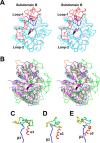
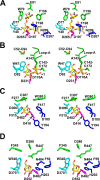
Arthrobacter globiformis T6 isomalto-dextranase (AgIMD) is an enzyme that liberates isomaltose from the non-reducing end of a polymer of glucose, dextran. AgIMD is classified as a member of the glycoside hydrolase family (GH) 27, which comprises mainly α-galactosidases and α-N-acetylgalactosaminidases, whereas AgIMD does not show α-galactosidase or α-N-acetylgalactosaminidase activities. Here, we determined the crystal structure of AgIMD. AgIMD consists of the following three domains: A, C, and D. Domains A and C are identified as a (β/α)8-barrel catalytic domain and an antiparallel β-structure, respectively, both of which are commonly found in GH27 enzymes. However, domain A of AgIMD has subdomain B, loop-1, and loop-2, all of which are not found in GH27 human α-galactosidase. AgIMD in a complex with trisaccharide panose shows that Asp-207, a residue in loop-1, is involved in subsite +1. Kinetic parameters of the wild-type and mutant enzymes for the small synthetic saccharide p-nitrophenyl α-isomaltoside and the polysaccharide dextran were compared, showing that Asp-207 is important for the catalysis of dextran. Domain D is classified as carbohydrate-binding module (CBM) 35, and an isomaltose molecule is seen in this domain in the AgIMD-isomaltose complex. Domain D is highly homologous to CBM35 domains found in GH31 and GH66 enzymes. The results here indicate that some features found in GH13, -31, and -66 enzymes, such as subdomain B, residues at the subsite +1, and the CBM35 domain, are also observed in the GH27 enzyme AgIMD and thus provide insights into the evolutionary relationships among GH13, -27, -31, -36, and -66 enzymes.
Keywords: carbohydrate metabolism; enzyme kinetics; glycoside hydrolase; phylogenetics; x-ray crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.Acta Crystallogr F Struct Biol Commun. 2016 Oct 1;72(Pt 10):750-761. doi: 10.1107/S2053230X16014072. Epub 2016 Sep 22. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 27710940 Free PMC article.
-
Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31.FEBS J. 2020 Mar;287(6):1195-1207. doi: 10.1111/febs.15074. Epub 2019 Oct 11. FEBS J. 2020. PMID: 31552702
-
Crystal structure of α-1,4-glucan lyase, a unique glycoside hydrolase family member with a novel catalytic mechanism.J Biol Chem. 2013 Sep 13;288(37):26764-74. doi: 10.1074/jbc.M113.485896. Epub 2013 Jul 31. J Biol Chem. 2013. PMID: 23902768 Free PMC article.
-
Structural and biochemical characterization of novel bacterial α-galactosidases belonging to glycoside hydrolase family 31.Biochem J. 2015 Jul 1;469(1):145-58. doi: 10.1042/BJ20150261. Epub 2015 May 5. Biochem J. 2015. PMID: 25942325
-
Structure-function analysis of bacterial GH31 α-galactosidases specific for α-(1→4)-galactobiose.FEBS J. 2023 Oct;290(20):4984-4998. doi: 10.1111/febs.16904. Epub 2023 Jul 19. FEBS J. 2023. PMID: 37438884
Cited by
-
Improving the thermostability of a GH97 dextran glucosidase by rational design.Biotechnol Lett. 2020 Nov;42(11):2211-2221. doi: 10.1007/s10529-020-02928-8. Epub 2020 Jun 1. Biotechnol Lett. 2020. PMID: 32488441
-
Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-β-1,3-galactanase from Phanerochaete chrysosporium.J Biol Chem. 2020 Dec 25;295(52):18539-18552. doi: 10.1074/jbc.RA120.016149. Epub 2020 Oct 22. J Biol Chem. 2020. PMID: 33093171 Free PMC article.
-
Review on recent advances in the properties, production and applications of microbial dextranases.World J Microbiol Biotechnol. 2023 Jul 4;39(9):242. doi: 10.1007/s11274-023-03691-4. World J Microbiol Biotechnol. 2023. PMID: 37400664 Review.
-
OsAPSE modulates non-covalent interactions between arabinogalactan protein O-glycans and pectin in rice cell walls.Front Plant Sci. 2025 May 22;16:1588802. doi: 10.3389/fpls.2025.1588802. eCollection 2025. Front Plant Sci. 2025. PMID: 40475901 Free PMC article.
-
Purification and characterization of cold-adapted and salt-tolerant dextranase from Cellulosimicrobium sp. THN1 and its potential application for treatment of dental plaque.Front Microbiol. 2022 Nov 11;13:1012957. doi: 10.3389/fmicb.2022.1012957. eCollection 2022. Front Microbiol. 2022. PMID: 36439846 Free PMC article.
References
-
- Sawai T., Toriyama K., and Yano K. (1974) A bacterial dextranase releasing only isomaltose from dextrans. J. Biochem. 75, 105–112 - PubMed
-
- Sawai T., Ohara S., Ichimi Y., Okaji S., Hisada K., and Fukaya N. (1981) Purification and some properties of the isomalto-dextranase of Actinomadura strain R10 and comparison with that of Arthrobacter globiformis T6. Carbohydr. Res. 89, 289–299
-
- Takayanagi T., Okada G., and Chiba S. (1987) Quantitative study of the anomeric forms of isomaltose and glucose produced by isomalto-dextranase and glucodextranase. Agric. Biol. Chem. 51, 2337–2341
-
- Nihira T, Mizuno M, Tonozuka T, Sakano Y, Mori T, and Okahata Y. (2005) Kinetic studies of site-directed mutational isomalto-dextranase-catalyzed hydrolytic reactions on a 27 MHz quartz-crystal microbalance. Biochemistry 44, 9456–9461 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

