Azide anions inhibit GH-18 endochitinase and GH-20 Exo β-N-acetylglucosaminidase from the marine bacterium Vibrio harveyi
- PMID: 26330565
- PMCID: PMC4892774
- DOI: 10.1093/jb/mvv087
Azide anions inhibit GH-18 endochitinase and GH-20 Exo β-N-acetylglucosaminidase from the marine bacterium Vibrio harveyi
Abstract
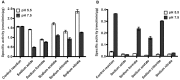
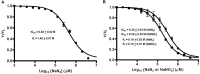
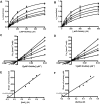
Vibrio harveyi is a bioluminescent marine bacterium that utilizes chitin as its sole source of energy. In the course of chitin degradation, the bacterium primarily secretes an endochitinase A (VhChiA) to hydrolyze chitin, generating chitooligosaccharide fragments that are readily transported into the cell and broken down to GlcNAc monomers by an exo β-N-acetylglucosaminidase (VhGlcNAcase). Here we report that sodium salts, especially sodium azide, inhibit two classes of these chitin-degrading enzymes (VhChiA and VhGlcNAcase) with distinct modes of action. Kinetic analysis of the enzymatic hydrolysis of pNP-glycoside substrates reveals that sodium azide inhibition of VhChiA has a mixed-type mode, but that it inhibits VhGlcNAcase competitively. We propose that azide anions inhibit chitinase activity by acting as strong nucleophiles that attack Cγ of the catalytic Glu or Cβ of the neighbouring Asp residues. Azide anions may bind not only to the catalytic centre, but also to the other subsites in the substrate-binding cleft of VhChiA. In contrast, azide anions may merely occupy the small-binding pocket of VhGlcNAcase, thereby blocking the accessibility of its active site by short-chain substrates.
Keywords: chitin turnover; family 18 chitinase; family 20 N-acetylglucosaminidase; reversible inhibition; sodium azide.
© The Authors 2015. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures





References
-
- Bassler B.L., Yu C., Lee Y.C., Roseman S. (1991) Chitin utilization by marine bacteria. J. Biol. Chem. 266, 24276–24286 - PubMed
-
- Cohen-Kupiec R., Chet I. (1998) The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9, 270–277 - PubMed
-
- Suginta W., Vongsuwan A., Songsiriritthigul C., Svasti J., Prinz H. (2005) Enzymatic properties of wild-type and active site mutants of chitinase A from Vibrio carchariae, as revealed by HPLC-MS. FEBS J. 272, 3376–3386 - PubMed
-
- Matsuo Y., Kurita M., Park J.K., Tanaka k., Nakagawa T., Kawamukai M., Matsuda H. (1999) Purification, characterization and gene analysis of N-acetylglucosaminidase from Enterobacter sp. G-1. Biosci. Biotechnol. Biochem . 63, 1261–1268 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

