Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators
- PMID: 26330747
- PMCID: PMC4551281
Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators
Abstract
Purpose: Gene expression correlates with local chromatin structure. Our studies have mapped histone post-translational modifications, RNA polymerase II (pol II), and transcription factor Pax6 in lens chromatin. These data represent the first genome-wide insights into the relationship between lens chromatin structure and lens transcriptomes and serve as an excellent source for additional data analysis and refinement. The principal lens proteins, the crystallins, are encoded by predominantly expressed mRNAs; however, the regulatory mechanisms underlying their high expression in the lens remain poorly understood.
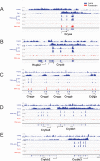
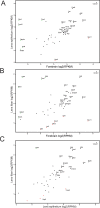
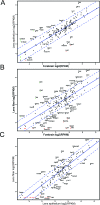
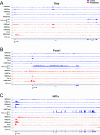
Methods: The formaldehyde-assisted identification of regulatory regions (FAIRE-Seq) was employed to analyze newborn lens chromatin. ChIP-seq and RNA-seq data published earlier (GSE66961) have been used to assist in FAIRE-seq data interpretation. RNA transcriptomes from murine lens epithelium, lens fibers, erythrocytes, forebrain, liver, neurons, and pancreas were compared to establish the gene expression levels of the most abundant mRNAs versus median gene expression across other differentiated cells.
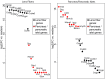
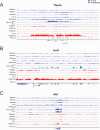
Results: Normalized RNA expression data from multiple tissues show that crystallins rank among the most highly expressed genes in mammalian cells. These findings correlate with the extremely high abundance of pol II all across the crystallin loci, including crystallin genes clustered on chromosomes 1 and 5, as well as within regions of "open" chromatin, as identified by FAIRE-seq. The expression levels of mRNAs encoding DNA-binding transcription factors (e.g., Foxe3, Hsf4, Maf, Pax6, Prox1, Sox1, and Tfap2a) revealed that their transcripts form "clusters" of abundant mRNAs in either lens fibers or lens epithelium. The expression of three autophagy regulatory mRNAs, encoding Tfeb, FoxO1, and Hif1α, was found within a group of lens preferentially expressed transcription factors compared to the E12.5 forebrain.
Conclusions: This study reveals novel features of lens chromatin, including the remarkably high abundance of pol II at the crystallin loci that exhibit features of "open" chromatin. Hsf4 ranks among the most abundant fiber cell-preferred DNA-binding transcription factors. Notable transcripts, including Atf4, Ctcf, E2F4, Hey1, Hmgb1, Mycn, RXRβ, Smad4, Sp1, and Taf1 (transcription factors) and Ctsd, Gabarapl1, and Park7 (autophagy regulators) have been identified with high levels of expression in lens fibers, which suggests specific roles in lens fiber cell terminal differentiation.
Figures




References
-
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

