Quantitative Analysis of Intracellular Fluorescent Foci in Live Bacteria
- PMID: 26331246
- PMCID: PMC4564679
- DOI: 10.1016/j.bpj.2015.07.013
Quantitative Analysis of Intracellular Fluorescent Foci in Live Bacteria
Abstract
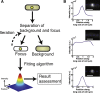
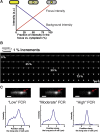
Fluorescence microscopy has revolutionized in vivo cellular biology. Through the specific labeling of a protein of interest with a fluorescent protein, one is able to study movement and colocalization, and even count individual proteins in a live cell. Different algorithms exist to quantify the total intensity and position of a fluorescent focus. Although these algorithms have been rigorously studied for in vitro conditions, which are greatly different than the in-homogenous and variable cellular environments, their exact limits and applicability in the context of a live cell have not been thoroughly and systematically evaluated. In this study, we quantitatively characterize the influence of different background subtraction algorithms on several focus analysis algorithms. We use, to our knowledge, a novel approach to assess the sensitivity of the focus analysis algorithms to background removal, in which simulated and experimental data are combined to maintain full control over the sensitivity of a focus within a realistic background of cellular fluorescence. We demonstrate that the choice of algorithm and the corresponding error are dependent on both the brightness of the focus, and the cellular context. Expectedly, focus intensity estimation and localization accuracy suffer in all algorithms at low focus to background ratios, with the bacteroidal background subtraction in combination with the median excess algorithm, and the region of interest background subtraction in combination with a two-dimensional Gaussian fit algorithm, performing the best. We furthermore show that the choice of background subtraction algorithm is dependent on the expression level of the protein under investigation, and that the localization error is dependent on the distance of a focus from the bacterial edge and pole. Our results establish a set of guidelines for what signals can be analyzed to give a targeted spatial and intensity accuracy within a bacterial cell.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

