Distribution of Pico- and Nanosecond Motions in Disordered Proteins from Nuclear Spin Relaxation
- PMID: 26331256
- PMCID: PMC4564687
- DOI: 10.1016/j.bpj.2015.06.069
Distribution of Pico- and Nanosecond Motions in Disordered Proteins from Nuclear Spin Relaxation
Abstract
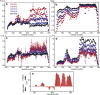
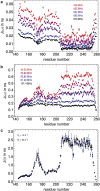
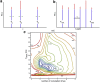
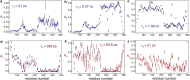
Intrinsically disordered proteins and intrinsically disordered regions (IDRs) are ubiquitous in the eukaryotic proteome. The description and understanding of their conformational properties require the development of new experimental, computational, and theoretical approaches. Here, we use nuclear spin relaxation to investigate the distribution of timescales of motions in an IDR from picoseconds to nanoseconds. Nitrogen-15 relaxation rates have been measured at five magnetic fields, ranging from 9.4 to 23.5 T (400-1000 MHz for protons). This exceptional wealth of data allowed us to map the spectral density function for the motions of backbone NH pairs in the partially disordered transcription factor Engrailed at 11 different frequencies. We introduce an approach called interpretation of motions by a projection onto an array of correlation times (IMPACT), which focuses on an array of six correlation times with intervals that are equidistant on a logarithmic scale between 21 ps and 21 ns. The distribution of motions in Engrailed varies smoothly along the protein sequence and is multimodal for most residues, with a prevalence of motions around 1 ns in the IDR. We show that IMPACT often provides better quantitative agreement with experimental data than conventional model-free or extended model-free analyses with two or three correlation times. We introduce a graphical representation that offers a convenient platform for a qualitative discussion of dynamics. Even when relaxation data are only acquired at three magnetic fields that are readily accessible, the IMPACT analysis gives a satisfactory characterization of spectral density functions, thus opening the way to a broad use of this approach.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. - PubMed
-
- Dunker A.K., Lawson J.D., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. - PubMed
-
- Jensen M.R., Markwick P.R.L., Blackledge M. Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure. 2009;17:1169–1185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

