The Complex Energy Landscape of the Protein IscU
- PMID: 26331259
- PMCID: PMC4564936
- DOI: 10.1016/j.bpj.2015.07.045
The Complex Energy Landscape of the Protein IscU
Abstract
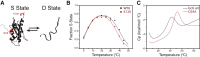
IscU, the scaffold protein for iron-sulfur (Fe-S) cluster biosynthesis in Escherichia coli, traverses a complex energy landscape during Fe-S cluster synthesis and transfer. Our previous studies showed that IscU populates two interconverting conformational states: one structured (S) and one largely disordered (D). Both states appear to be functionally important because proteins involved in the assembly or transfer of Fe-S clusters have been shown to interact preferentially with either the S or D state of IscU. To characterize the complex structure-energy landscape of IscU, we employed NMR spectroscopy, small-angle x-ray scattering (SAXS), and differential scanning calorimetry. Results obtained for IscU at pH 8.0 show that its S state is maximally populated at 25°C and that heating or cooling converts the protein toward the D state. Results from NMR and DSC indicate that both the heat- and cold-induced S→D transitions are cooperative and two-state. Low-resolution structural information from NMR and SAXS suggests that the structures of the cold-induced and heat-induced D states are similar. Both states exhibit similar (1)H-(15)N HSQC spectra and the same pattern of peptidyl-prolyl peptide bond configurations by NMR, and both appear to be similarly expanded compared with the S state based on analysis of SAXS data. Whereas in other proteins the cold-denatured states have been found to be slightly more compact than the heat-denatured states, these two states occupy similar volumes in IscU.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery.FEBS Lett. 2013 Apr 17;587(8):1172-9. doi: 10.1016/j.febslet.2013.01.003. Epub 2013 Jan 16. FEBS Lett. 2013. PMID: 23333622 Free PMC article. Review.
-
Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase.Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):454-9. doi: 10.1073/pnas.1114372109. Epub 2011 Dec 27. Proc Natl Acad Sci U S A. 2012. PMID: 22203963 Free PMC article.
-
Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU).J Biol Chem. 2013 Oct 4;288(40):28755-70. doi: 10.1074/jbc.M113.482042. Epub 2013 Aug 12. J Biol Chem. 2013. PMID: 23940031 Free PMC article.
-
Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones.Biochemistry. 2008 Dec 2;47(48):12795-801. doi: 10.1021/bi801565j. Biochemistry. 2008. PMID: 18986169
-
Tangled web of interactions among proteins involved in iron-sulfur cluster assembly as unraveled by NMR, SAXS, chemical crosslinking, and functional studies.Biochim Biophys Acta. 2015 Jun;1853(6):1416-28. doi: 10.1016/j.bbamcr.2014.11.020. Epub 2014 Nov 22. Biochim Biophys Acta. 2015. PMID: 25450980 Free PMC article. Review.
Cited by
-
Light from a firefly at temperatures considerably higher and lower than normal.Sci Rep. 2021 Jun 14;11(1):12498. doi: 10.1038/s41598-021-91839-3. Sci Rep. 2021. PMID: 34127729 Free PMC article.
-
Biomolecular NMR: Past and future.Arch Biochem Biophys. 2017 Aug 15;628:3-16. doi: 10.1016/j.abb.2017.05.003. Epub 2017 May 8. Arch Biochem Biophys. 2017. PMID: 28495511 Free PMC article.
-
New Techniques for Ancient Proteins: Direct Coupling Analysis Applied on Proteins Involved in Iron Sulfur Cluster Biogenesis.Front Mol Biosci. 2017 Jun 15;4:40. doi: 10.3389/fmolb.2017.00040. eCollection 2017. Front Mol Biosci. 2017. PMID: 28664160 Free PMC article.
-
From Protein Design to the Energy Landscape of a Cold Unfolding Protein.J Phys Chem B. 2022 Feb 17;126(6):1212-1231. doi: 10.1021/acs.jpcb.1c10750. Epub 2022 Feb 7. J Phys Chem B. 2022. PMID: 35128921 Free PMC article.
-
Mechanistic Insights into IscU Conformation Regulation for Fe-S Cluster Biogenesis Revealed by Variable Temperature Electrospray Ionization Native Ion Mobility Mass Spectrometry.Biochemistry. 2022 Dec 6;61(23):2733-2741. doi: 10.1021/acs.biochem.2c00429. Epub 2022 Nov 9. Biochemistry. 2022. PMID: 36351081 Free PMC article.
References
-
- Privalov P.L. Cold denaturation of proteins. Crit. Rev. Biochem. Mol. Biol. 1990;25:281–305. - PubMed
-
- Wong K.B., Freund S.M., Fersht A.R. Cold denaturation of barstar: 1H, 15N and 13C NMR assignment and characterisation of residual structure. J. Mol. Biol. 1996;259:805–818. - PubMed
-
- Babu C.R., Hilser V.J., Wand A.J. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat. Struct. Mol. Biol. 2004;11:352–357. - PubMed
-
- Van Horn W.D., Simorellis A.K., Flynn P.F. Low-temperature studies of encapsulated proteins. J. Am. Chem. Soc. 2005;127:13553–13560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

