Hirsutinolide Series Inhibit Stat3 Activity, Alter GCN1, MAP1B, Hsp105, G6PD, Vimentin, TrxR1, and Importin α-2 Expression, and Induce Antitumor Effects against Human Glioma
- PMID: 26331426
- PMCID: PMC4834983
- DOI: 10.1021/acs.jmedchem.5b00686
Hirsutinolide Series Inhibit Stat3 Activity, Alter GCN1, MAP1B, Hsp105, G6PD, Vimentin, TrxR1, and Importin α-2 Expression, and Induce Antitumor Effects against Human Glioma
Abstract
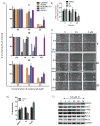
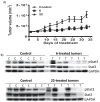
We report that hirsutinolide series, 6, 7, 10, 11, 20, and 22, and the semisynthetic analogues, 30, 31, 33, and 36, inhibit constitutively active signal transducer and activator of transcription (Stat)3 and malignant glioma phenotype. A position 13 lipophilic ester group is required for activity. Molecular modeling and nuclear magnetic resonance structural analyses reveal direct hirsutinolide:Stat3 binding. One-hour treatment of cells with 6 and 22 also upregulated importin subunit α-2 levels and repressed translational activator GCN1, microtubule-associated protein (MAP)1B, thioredoxin reductase (TrxR)1 cytoplasmic isoform 3, glucose-6-phosphate 1-dehydrogenase isoform a, Hsp105, vimentin, and tumor necrosis factor α-induced protein (TNAP)2 expression. Active hirsutinolides inhibited anchorage-dependent and three-dimensional spheroid growth, survival, and migration of human glioma lines and glioma patients' tumor-derived xenograft cells harboring constitutively active Stat3. Oral gavage delivery of 6 or 22 inhibited human glioma tumor growth in subcutaneous mouse xenografts. The inhibition of Stat3 signaling represents part of the hirsutinolide-mediated mechanisms to induce antitumor effects.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical
Miscellaneous

