Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers
- PMID: 26332016
- PMCID: PMC4986832
- DOI: 10.1039/c5an00688k
Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers
Abstract
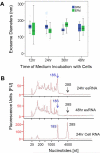
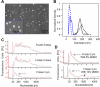
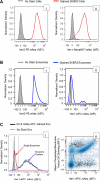
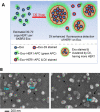
As a type of secreted membrane vesicle, exosomes are an emerging mode of cell-to-cell communication. Yet as exosome samples are commonly contaminated with other extracellular vesicles, the biological roles of exosomes in regulating immunity and promoting oncogenesis remain controversial. Wondering whether existing methods could distort our view of exosome biology, we compared two direct methods for imaging extracellular vesicles and quantified the impact of different production and storage conditions on the quality of exosome samples. Scanning electron microscopy (SEM) was compared to transmission electron microscopy (TEM) as alternatives to examine the morphology of exosomes. Using SEM, we were able to distinguish exosomes from other contaminating extracellular vesicles based on the size distribution. More importantly, freezing of samples prior to SEM imaging made it more difficult to distinguish exosomes from extracellular vesicles secreted during cell death. In addition to morphology, the quality of RNA contained within the exosomes was characterized under different storage conditions, where freezing of samples also degraded RNA. Finally, we developed a new flow cytometry approach to assay transmembrane proteins on exosomes. While high-copy-number proteins could be readily detected, detecting low-copy-number proteins was improved using a lipophilic tracer that clustered exosomes. To illustrate this, we observed that exosomes derived from SKBR3 cells, a cell model for human HER2+ breast cancer, contained both HER1 and HER2 but at different levels of abundance. Collectively, these new methods will help to ensure a consistent framework to identify specific roles that exosomes play in regulating cell-to-cell communication.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

