Yeast Mitochondrial Transcription Factor Mtf1 Determines the Precision of Promoter-Directed Initiation of RNA Polymerase Rpo41
- PMID: 26332125
- PMCID: PMC4558008
- DOI: 10.1371/journal.pone.0136879
Yeast Mitochondrial Transcription Factor Mtf1 Determines the Precision of Promoter-Directed Initiation of RNA Polymerase Rpo41
Abstract
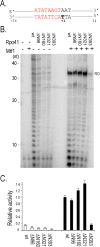
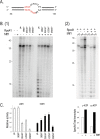
Despite their clear T7-bacteriophage origin, mitochondrial RNA polymerases have evolved to require transcription factors. All mitochondrial polymerases contain an extra N-terminal domain that has no counterpart in the self-proficient phage enzyme, which is therefore hypothesized to interact with transcription factors. We studied a series of N-terminal deletion mutants of yeast mitochondrial RNA polymerase, Rpo41, and have found that the N-terminal region does not abolish the effects of Mtf1; rather it contributes directly to enzyme catalysis. Mtf1 can rescue the defective Rpo41 enzymes resulted from N-terminal domain deletions. Although Rpo41 appears to have retained all promoter recognition elements found in T7 RNAP, the elements are not independently functional, and Mtf1 is necessary and sufficient for holoenzyme promoter-directed transcription activity.
Conflict of interest statement
Figures




Similar articles
-
The N-terminal domain of the yeast mitochondrial RNA polymerase regulates multiple steps of transcription.J Biol Chem. 2011 May 6;286(18):16109-20. doi: 10.1074/jbc.M111.228023. Epub 2011 Mar 18. J Biol Chem. 2011. PMID: 21454631 Free PMC article.
-
Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation.J Biol Chem. 2010 Feb 5;285(6):3949-3956. doi: 10.1074/jbc.M109.050732. Epub 2009 Dec 11. J Biol Chem. 2010. PMID: 20008320 Free PMC article.
-
Expression and purification of wild type and mutant forms of the yeast mitochondrial core RNA polymerase, Rpo41.Protein Expr Purif. 2004 May;35(1):126-30. doi: 10.1016/j.pep.2003.12.022. Protein Expr Purif. 2004. PMID: 15039075
-
Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):930-8. doi: 10.1016/j.bbagrm.2012.02.003. Epub 2012 Feb 14. Biochim Biophys Acta. 2012. PMID: 22353467 Free PMC article. Review.
-
Mitochondrial transcription: is a pattern emerging?Mol Microbiol. 1993 Apr;8(1):1-4. doi: 10.1111/j.1365-2958.1993.tb01197.x. Mol Microbiol. 1993. PMID: 8497187 Review.
Cited by
-
The Identification of the Mitochondrial DNA Polymerase γ (Mip1) of the Entomopathogenic Fungus Metarhizium brunneum.Microorganisms. 2024 May 23;12(6):1052. doi: 10.3390/microorganisms12061052. Microorganisms. 2024. PMID: 38930434 Free PMC article.
-
High yields and soluble expression of superoxide dismutases in Escherichia coli due to the HIV-1 Tat peptide via increases in mRNA transcription.Exp Mol Med. 2016 Oct 14;48(10):e264. doi: 10.1038/emm.2016.91. Exp Mol Med. 2016. PMID: 27741225 Free PMC article.
-
The Yeast Mitochondrial RNA Polymerase and Transcription Factor Complex Catalyzes Efficient Priming of DNA Synthesis on Single-stranded DNA.J Biol Chem. 2016 Aug 5;291(32):16828-39. doi: 10.1074/jbc.M116.740282. Epub 2016 Jun 16. J Biol Chem. 2016. PMID: 27311715 Free PMC article.
-
Orphan proteins of unknown function in the mitochondrial intermembrane space proteome: New pathways and metabolic cross-talk.Biochim Biophys Acta. 2016 Nov;1863(11):2613-2623. doi: 10.1016/j.bbamcr.2016.07.004. Epub 2016 Jul 15. Biochim Biophys Acta. 2016. PMID: 27425144 Free PMC article. Review.
-
The Pet127 protein is a mitochondrial 5'-to-3' exoribonuclease from the PD-(D/E)XK superfamily involved in RNA maturation and intron degradation in yeasts.RNA. 2022 May;28(5):711-728. doi: 10.1261/rna.079083.121. Epub 2022 Feb 23. RNA. 2022. PMID: 35197365 Free PMC article.
References
-
- Fuste J. M., Wanrooij S., Jemt E., Granycome C. E., Cluett T. J., Shi Y., et al. (2009). Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell 37, 67–78. - PubMed
-
- Wilcoxen S. E., Peterson C. R., Winkley C. S., Keller M. J. & Jaehning J. A. (1988). Two forms of RPO41-dependent RNA polymerase. Regulation of the RNA polymerase by glucose repression may control yeast mitochondrial gene expression. J Biol Chem 263, 12346–51. - PubMed
-
- Schinkel A. H., Groot Koerkamp, M. J., Van der Horst G. T., Touw E. P., Osinga K. A., Van der Bliek A. M., et al. (1986). Characterization of the promoter of the large ribosomal RNA gene in yeast mitochondria and separation of mitochondrial RNA polymerase into two different functional components. EMBO J 5, 1041–7. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

