What are the Evolutionary Origins of Mitochondria? A Complex Network Approach
- PMID: 26332127
- PMCID: PMC4557972
- DOI: 10.1371/journal.pone.0134988
What are the Evolutionary Origins of Mitochondria? A Complex Network Approach
Abstract
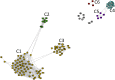
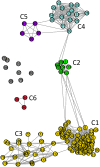
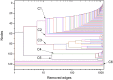
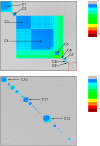
Mitochondria originated endosymbiotically from an Alphaproteobacteria-like ancestor. However, it is still uncertain which extant group of Alphaproteobacteria is phylogenetically closer to the mitochondrial ancestor. The proposed groups comprise the order Rickettsiales, the family Rhodospirillaceae, and the genus Rickettsia. In this study, we apply a new complex network approach to investigate the evolutionary origins of mitochondria, analyzing protein sequences modules in a critical network obtained through a critical similarity threshold between the studied sequences. The dataset included three ATP synthase subunits (4, 6, and 9) and its alphaproteobacterial homologs (b, a, and c). In all the subunits, the results gave no support to the hypothesis that Rickettsiales are closely related to the mitochondrial ancestor. Our findings support the hypothesis that mitochondria share a common ancestor with a clade containing all Alphaproteobacteria orders, except Rickettsiales.
Conflict of interest statement
Figures
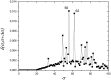



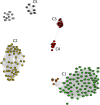
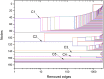
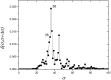


References
-
- Atteia A, Adrait A, Brugière S, Tardif M, van Lis R, Deusch O, et al. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-Proteobacterial mitochondrial ancestor. Molecular Biology and Evolution 26:1533–1548. 10.1093/molbev/msp068 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

