Methyl Relaxation Measurements Reveal Patterns of Fast Dynamics in a Viral RNA-Directed RNA Polymerase
- PMID: 26333183
- PMCID: PMC5259806
- DOI: 10.1021/acs.biochem.5b00828
Methyl Relaxation Measurements Reveal Patterns of Fast Dynamics in a Viral RNA-Directed RNA Polymerase
Abstract
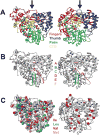
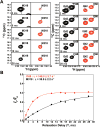
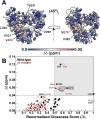
Molecular dynamics (MD) simulations combined with biochemical studies have suggested the presence of long-range networks of functionally relevant conformational flexibility on the nanosecond time scale in single-subunit RNA polymerases in many RNA viruses. However, experimental verification of these dynamics at a sufficient level of detail has been lacking. Here we describe the fast, picosecond to nanosecond dynamics of an archetypal viral RNA-directed RNA polymerase (RdRp), the 75 kDa P2 protein from cystovirus ϕ12, using analyses of (1)H-(1)H dipole-dipole cross-correlated relaxation at the methyl positions of Ile (δ1), Leu, Val, and Met residues. Our results, which represent the most detailed experimental characterization of fast dynamics in a viral RdRp until date, reveal a highly connected dynamic network as predicted by MD simulations of related systems. Our results suggest that the entry portals for template RNA and substrate NTPs are relatively disordered, while conserved motifs involved in metal binding, nucleotide selection, and catalysis display greater rigidity. Perturbations at the active site through metal binding or functional mutation affect dynamics not only in the immediate vicinity but also at remote regions. Comparison with the limited experimental and extensive functional and in silico results available for homologous systems suggests conservation of the overall pattern of dynamics in viral RdRps.
Figures


References
-
- Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. - PubMed
-
- Ontoria JM, Rydberg EH, Di Marco S, Tomei L, Attenni B, Malancona S, Martin Hernando JI, Gennari N, Koch U, Narjes F, Rowley M, Summa V, Carroll SS, Olsen DB, De Francesco R, Altamura S, Migliaccio G, Carfí A. Identification and biological evaluation of a series of 1H-benzo[de]isoquinoline-1,3(2H)-diones as hepatitis C virus NS5B polymerase inhibitors. J Med Chem. 2009;52:5217–5227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

