Regulation of hedgehog Ligand Expression by the N-End Rule Ubiquitin-Protein Ligase Hyperplastic Discs and the Drosophila GSK3β Homologue, Shaggy
- PMID: 26334301
- PMCID: PMC4559392
- DOI: 10.1371/journal.pone.0136760
Regulation of hedgehog Ligand Expression by the N-End Rule Ubiquitin-Protein Ligase Hyperplastic Discs and the Drosophila GSK3β Homologue, Shaggy
Abstract
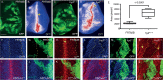
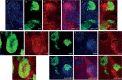
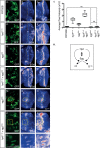
Hedgehog (Hh) morphogen signalling plays an essential role in tissue development and homeostasis. While much is known about the Hh signal transduction pathway, far less is known about the molecules that regulate the expression of the hedgehog (hh) ligand itself. Here we reveal that Shaggy (Sgg), the Drosophila melanogaster orthologue of GSK3β, and the N-end Rule Ubiquitin-protein ligase Hyperplastic Discs (Hyd) act together to co-ordinate Hedgehog signalling through regulating hh ligand expression and Cubitus interruptus (Ci) expression. Increased hh and Ci expression within hyd mutant clones was effectively suppressed by sgg RNAi, placing sgg downstream of hyd. Functionally, sgg RNAi also rescued the adult hyd mutant head phenotype. Consistent with the genetic interactions, we found Hyd to physically interact with Sgg and Ci. Taken together we propose that Hyd and Sgg function to co-ordinate hh ligand and Ci expression, which in turn influences important developmental signalling pathways during imaginal disc development. These findings are important as tight temporal/spatial regulation of hh ligand expression underlies its important roles in animal development and tissue homeostasis. When deregulated, hh ligand family misexpression underlies numerous human diseases (e.g., colorectal, lung, pancreatic and haematological cancers) and developmental defects (e.g., cyclopia and polydactyly). In summary, our Drosophila-based findings highlight an apical role for Hyd and Sgg in initiating Hedgehog signalling, which could also be evolutionarily conserved in mammals.
Conflict of interest statement
Figures





References
-
- Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108(6):823–35. Epub 2002/04/17. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/H012869/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 2P40OD010949-10A1/OD/NIH HHS/United States
- P40 OD010949/OD/NIH HHS/United States
- BB/H0128691/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- RA1734/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

