Exposure to fluorescent light triggers down regulation of genes involved with mitotic progression in Xiphophorus skin
- PMID: 26334372
- PMCID: PMC4662871
- DOI: 10.1016/j.cbpc.2015.08.006
Exposure to fluorescent light triggers down regulation of genes involved with mitotic progression in Xiphophorus skin
Abstract
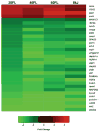
We report RNA-Seq results from skin of X. maculatus Jp 163 B after exposure to various doses of "cool white" fluorescent light (FL). We show that FL exposure incites a genetic transcriptional response in skin nearly as great as observed for UVB exposure; however, the gene sets modulated due to exposure to the two light sources are quite different. Known light responsive genes involved in maintaining circadian cycling (e.g., clock, cry2a, cry1b, per1b, per2, per3, and arntl1a) exhibited expected shifts in transcriptional expression upon FL exposure. Exposure to FL also resulted in down-regulated transcription of many genes involved with cell cycle progression (e.g., cdc20, cdc45, cdca7b, plk1, cdk1, ccnb-3, and cdca7a) and chromosome segregation (e.g., cenpe, cenpf, cenpi, cenpk, cenpo, cenpp, and cenpu; cep70; knstrm, kntc, mcm2, mcm5; smc2). In addition, several DNA replication and recombination repair genes (e.g., pola1, pole, rec52, rad54l, rpa1, and parpbp) exhibit reduced expression in FL exposed X. maculatus skin. Some genes down modulated by FL are known to be associated with DNA repair and human diseases (e.g., atm2, brip1, fanc1, fancl, and xrcc4). The overall suppression of genes involved with mitotic progression in the skin of adult fish is consistent with entry into the light phase of the circadian cycle. Current efforts are aimed at determining specific wavelengths that may lead to differential expression among the many genes affected by fluorescent light exposure.
Keywords: Cell cycle progression; Circadian; Cool white light; DNA damage response; Differential gene expression; Fluorescent light; RNA-Seq.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Waveband specific transcriptional control of select genetic pathways in vertebrate skin (Xiphophorus maculatus).BMC Genomics. 2018 May 10;19(1):355. doi: 10.1186/s12864-018-4735-5. BMC Genomics. 2018. PMID: 29747585 Free PMC article.
-
Exposure to 4100K fluorescent light elicits sex specific transcriptional responses in Xiphophorus maculatus skin.Comp Biochem Physiol C Toxicol Pharmacol. 2018 Jun;208:96-104. doi: 10.1016/j.cbpc.2017.09.008. Epub 2017 Sep 29. Comp Biochem Physiol C Toxicol Pharmacol. 2018. PMID: 28965926 Free PMC article.
-
Molecular genetic response to varied wavelengths of light in Xiphophorus maculatus skin.Comp Biochem Physiol C Toxicol Pharmacol. 2015 Dec;178:104-115. doi: 10.1016/j.cbpc.2015.10.002. Epub 2015 Oct 14. Comp Biochem Physiol C Toxicol Pharmacol. 2015. PMID: 26460196 Free PMC article.
-
Mitotic recombination in yeast: elements controlling its incidence.Yeast. 2000 Jun 15;16(8):731-54. doi: 10.1002/1097-0061(20000615)16:8<731::AID-YEA586>3.0.CO;2-L. Yeast. 2000. PMID: 10861900 Review.
-
Schizosaccharomyces pombe Assays to Study Mitotic Recombination Outcomes.Genes (Basel). 2020 Jan 10;11(1):79. doi: 10.3390/genes11010079. Genes (Basel). 2020. PMID: 31936815 Free PMC article. Review.
Cited by
-
Common and distinct features of potentially predictive biomarkers in small cell lung carcinoma and large cell neuroendocrine carcinoma of the lung by systematic and integrated analysis.Mol Genet Genomic Med. 2020 Mar;8(3):e1126. doi: 10.1002/mgg3.1126. Epub 2020 Jan 25. Mol Genet Genomic Med. 2020. PMID: 31981472 Free PMC article.
-
Fluorescent Light Incites a Conserved Immune and Inflammatory Genetic Response within Vertebrate Organs (Danio Rerio, Oryzias Latipes and Mus Musculus).Genes (Basel). 2019 Apr 3;10(4):271. doi: 10.3390/genes10040271. Genes (Basel). 2019. PMID: 30987199 Free PMC article.
-
Fluorescent light exposure incites acute and prolonged immune responses in zebrafish (Danio rerio) skin.Comp Biochem Physiol C Toxicol Pharmacol. 2018 Jun;208:87-95. doi: 10.1016/j.cbpc.2017.09.009. Epub 2017 Sep 29. Comp Biochem Physiol C Toxicol Pharmacol. 2018. PMID: 28965927 Free PMC article.
-
Application of the Transcriptional Disease Signature (TDSs) to Screen Melanoma-Effective Compounds in a Small Fish Model.Sci Rep. 2019 Jan 24;9(1):530. doi: 10.1038/s41598-018-36656-x. Sci Rep. 2019. PMID: 30679619 Free PMC article.
-
Waveband specific transcriptional control of select genetic pathways in vertebrate skin (Xiphophorus maculatus).BMC Genomics. 2018 May 10;19(1):355. doi: 10.1186/s12864-018-4735-5. BMC Genomics. 2018. PMID: 29747585 Free PMC article.
References
-
- Ahmed FE, Setlow RB. Ultraviolet radiation-induced DNA damage and its photorepair in the skin of the platyfish Xiphophorus. Cancer Res. 1993;15(53):2249–55. - PubMed
-
- Armstrong TN, Reimschuessel R, Bradley BP. DNA damage, histological changes and DNA repair in larval Japanese medaka (Oryzias latipes) exposed to ultraviolet-B radiation. Aquat Toxicol. 2002;58 (1–2):1–14. - PubMed
-
- Bjarnason G, Jordan R. Circadian variation of cell proliferation and cell cycle protein expression in man: Clinical implications. Prog Cell Cycle Res. 2000;4:193–206. - PubMed
-
- Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “Circadian” Cycle. Cell Cycle. 2009;8 (6):8332–837. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

