Click chemistry armed enzyme-linked immunosorbent assay to measure palmitoylation by hedgehog acyltransferase
- PMID: 26334609
- PMCID: PMC4615133
- DOI: 10.1016/j.ab.2015.08.025
Click chemistry armed enzyme-linked immunosorbent assay to measure palmitoylation by hedgehog acyltransferase
Abstract
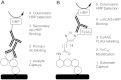
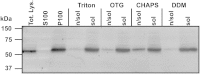
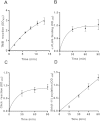
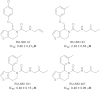
Hedgehog signaling is critical for correct embryogenesis and tissue development. However, on maturation, signaling is also found to be aberrantly activated in many cancers. Palmitoylation of the secreted signaling protein sonic hedgehog (Shh) by the enzyme hedgehog acyltransferase (Hhat) is required for functional signaling. To quantify this important posttranslational modification, many in vitro Shh palmitoylation assays employ radiolabeled fatty acids, which have limitations in terms of cost and safety. Here we present a click chemistry armed enzyme-linked immunosorbent assay (click-ELISA) for assessment of Hhat activity through acylation of biotinylated Shh peptide with an alkyne-tagged palmitoyl-CoA (coenzyme A) analogue. Click chemistry functionalization of the alkyne tag with azido-FLAG peptide allows analysis through an ELISA protocol and colorimetric readout. This assay format identified the detergent n-dodecyl β-d-maltopyranoside as an improved solubilizing agent for Hhat activity. Quantification of the potency of RU-SKI small molecule Hhat inhibitors by click-ELISA indicated IC50 values in the low- or sub-micromolar range. A stopped assay format was also employed that allows measurement of Hhat kinetic parameters where saturating substrate concentrations exceed the binding capacity of the streptavidin-coated plate. Therefore, click-ELISA represents a nonradioactive method for assessing protein palmitoylation in vitro that is readily expandable to other classes of protein lipidation.
Keywords: Click chemistry; Hedgehog acyltransferase; MBOAT; Protein palmitoylation.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Microfluidic Mobility Shift Assay for Real-Time Analysis of Peptide N-Palmitoylation.SLAS Discov. 2017 Apr;22(4):418-424. doi: 10.1177/2472555216689529. Epub 2017 Jan 31. SLAS Discov. 2017. PMID: 28296537 Free PMC article.
-
Palmitoylation of Hedgehog proteins by Hedgehog acyltransferase: roles in signalling and disease.Open Biol. 2021 Mar;11(3):200414. doi: 10.1098/rsob.200414. Epub 2021 Mar 3. Open Biol. 2021. PMID: 33653085 Free PMC article. Review.
-
In Vitro Analysis of Hedgehog Acyltransferase and Porcupine Fatty Acyltransferase Activities.Methods Mol Biol. 2019;2009:243-255. doi: 10.1007/978-1-4939-9532-5_19. Methods Mol Biol. 2019. PMID: 31152409 Free PMC article.
-
Hedgehog Acyltransferase Promotes Uptake of Palmitoyl-CoA across the Endoplasmic Reticulum Membrane.Cell Rep. 2019 Dec 24;29(13):4608-4619.e4. doi: 10.1016/j.celrep.2019.11.110. Cell Rep. 2019. PMID: 31875564 Free PMC article.
-
Palmitoylation of Hedgehog proteins.Vitam Horm. 2012;88:229-52. doi: 10.1016/B978-0-12-394622-5.00010-9. Vitam Horm. 2012. PMID: 22391306 Free PMC article. Review.
Cited by
-
Design, Synthesis, and Evaluation of Inhibitors of Hedgehog Acyltransferase.J Med Chem. 2024 Jan 25;67(2):1061-1078. doi: 10.1021/acs.jmedchem.3c01363. Epub 2024 Jan 10. J Med Chem. 2024. PMID: 38198226 Free PMC article.
-
Characterization of Hedgehog Acyltransferase Inhibitors Identifies a Small Molecule Probe for Hedgehog Signaling by Cancer Cells.ACS Chem Biol. 2016 Dec 16;11(12):3256-3262. doi: 10.1021/acschembio.6b00896. Epub 2016 Oct 25. ACS Chem Biol. 2016. PMID: 27779865 Free PMC article.
-
Synthesis and characterisation of 5-acyl-6,7-dihydrothieno[3,2-c]pyridine inhibitors of Hedgehog acyltransferase.Data Brief. 2016 Feb 10;7:257-81. doi: 10.1016/j.dib.2016.02.012. eCollection 2016 Jun. Data Brief. 2016. PMID: 27077078 Free PMC article.
-
Acylation-coupled lipophilic induction of polarisation (Acyl-cLIP): a universal assay for lipid transferase and hydrolase enzymes.Chem Sci. 2019 Jul 1;10(39):8995-9000. doi: 10.1039/c9sc01785b. eCollection 2019 Oct 21. Chem Sci. 2019. PMID: 31762980 Free PMC article.
-
cat-ELCCA: catalyzing drug discovery through click chemistry.Chem Commun (Camb). 2018 Jun 19;54(50):6531-6539. doi: 10.1039/c8cc02332h. Chem Commun (Camb). 2018. PMID: 29781014 Free PMC article.
References
-
- Chavda B., Arnott J.A., Planey S.L. Targeting protein palmitoylation: selective inhibitors and implications in disease. Expert Opin. Drug Discov. 2014;9:1005–1019. - PubMed
-
- Broncel M., Serwa R.A., Ciepla P., Krause E., Dallman M.J., Magee A.I. Multifunctional reagents for quantitative proteome-wide analysis of protein modification in human cells and dynamic profiling of protein lipidation during vertebrate development. Angew. Chem. Int. Ed. 2015;54:5948–5951. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

