Large-scale determination of previously unsolved protein structures using evolutionary information
- PMID: 26335199
- PMCID: PMC4602095
- DOI: 10.7554/eLife.09248
Large-scale determination of previously unsolved protein structures using evolutionary information
Abstract
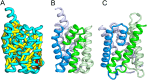
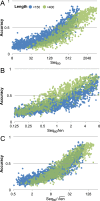
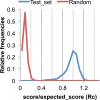
The prediction of the structures of proteins without detectable sequence similarity to any protein of known structure remains an outstanding scientific challenge. Here we report significant progress in this area. We first describe de novo blind structure predictions of unprecendented accuracy we made for two proteins in large families in the recent CASP11 blind test of protein structure prediction methods by incorporating residue-residue co-evolution information in the Rosetta structure prediction program. We then describe the use of this method to generate structure models for 58 of the 121 large protein families in prokaryotes for which three-dimensional structures are not available. These models, which are posted online for public access, provide structural information for the over 400,000 proteins belonging to the 58 families and suggest hypotheses about mechanism for the subset for which the function is known, and hypotheses about function for the remainder.
Keywords: B. subtilis; E. coli; H. salinarum; S. solfataricus; biophysics; co-evolution; evolutionary biology; genomics; protein family; protein fold; structural biology; structure prediction.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

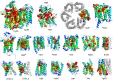


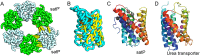





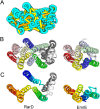
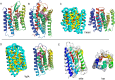
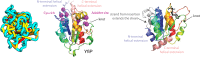


Similar articles
-
Improved de novo structure prediction in CASP11 by incorporating coevolution information into Rosetta.Proteins. 2016 Sep;84 Suppl 1(Suppl 1):67-75. doi: 10.1002/prot.24974. Epub 2016 Feb 24. Proteins. 2016. PMID: 26677056 Free PMC article.
-
Rosetta predictions in CASP5: successes, failures, and prospects for complete automation.Proteins. 2003;53 Suppl 6:457-68. doi: 10.1002/prot.10552. Proteins. 2003. PMID: 14579334
-
Protein structure determination using metagenome sequence data.Science. 2017 Jan 20;355(6322):294-298. doi: 10.1126/science.aah4043. Science. 2017. PMID: 28104891 Free PMC article.
-
Understanding the cell in terms of structure and function: insights from structural genomics.Curr Opin Biotechnol. 2006 Oct;17(5):457-64. doi: 10.1016/j.copbio.2006.07.004. Epub 2006 Aug 4. Curr Opin Biotechnol. 2006. PMID: 16890423 Review.
-
A tour of structural genomics.Nat Rev Genet. 2001 Oct;2(10):801-9. doi: 10.1038/35093574. Nat Rev Genet. 2001. PMID: 11584296 Review.
Cited by
-
Genetic analysis of the septal peptidoglycan synthase FtsWI complex supports a conserved activation mechanism for SEDS-bPBP complexes.PLoS Genet. 2021 Apr 15;17(4):e1009366. doi: 10.1371/journal.pgen.1009366. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33857142 Free PMC article.
-
Contingency and Entrenchment of Drug-Resistance Mutations in HIV Viral Proteins.J Phys Chem B. 2022 Dec 22;126(50):10622-10636. doi: 10.1021/acs.jpcb.2c06123. Epub 2022 Dec 9. J Phys Chem B. 2022. PMID: 36493468 Free PMC article.
-
Detection and sequence/structure mapping of biophysical constraints to protein variation in saturated mutational libraries and protein sequence alignments with a dedicated server.BMC Bioinformatics. 2016 Jun 17;17(1):242. doi: 10.1186/s12859-016-1124-4. BMC Bioinformatics. 2016. PMID: 27315797 Free PMC article.
-
Chemical tools to characterize peptidoglycan synthases.Curr Opin Chem Biol. 2019 Dec;53:44-50. doi: 10.1016/j.cbpa.2019.07.009. Epub 2019 Aug 26. Curr Opin Chem Biol. 2019. PMID: 31466035 Free PMC article. Review.
-
Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.Nat Commun. 2018 Mar 14;9(1):1078. doi: 10.1038/s41467-018-03477-5. Nat Commun. 2018. PMID: 29540682 Free PMC article.
References
-
- Abriata LA. An homology-and coevolution-consistent structural model of bacterial copper-tolerance protein CopM supports function as a ‘metal sponge’ and suggests regions for metal-dependent interactions with other proteins. 2015 bioRxiv:013581.
-
- Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. Journal of Molecular Microbiology and Biotechnology. 2002;4:71–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

