CYP2D6 Haplotype Determination Using Long Range Allele-Specific Amplification: Resolution of a Complex Genotype and a Discordant Genotype Involving the CYP2D6*59 Allele
- PMID: 26335396
- PMCID: PMC4630174
- DOI: 10.1016/j.jmoldx.2015.06.007
CYP2D6 Haplotype Determination Using Long Range Allele-Specific Amplification: Resolution of a Complex Genotype and a Discordant Genotype Involving the CYP2D6*59 Allele
Abstract
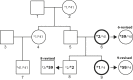
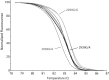
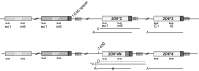
Cytochrome P450 (CYP) 2D6, a major contributor to the metabolism and bioactivation of many clinically used drugs, is encoded by a complex, highly polymorphic gene locus. To aid in the characterization of CYP2D6 allelic variation, we developed allele-specific long-range PCR (ASXL-PCR) to amplify only the allele of interest for further characterization by PCR. This development was achieved utilizing single-nucleotide polymorphisms in the upstream region of CYP2D6 and a universal CYP2D6-specific reverse primer. This approach was assessed and optimized on samples with known genotypes. The application of ASXL-PCR clarified a case with a complex genotype (CYP2D6*2x2/*4N+*4) by amplifying the duplicated gene units separately for subsequent analysis. Furthermore, ASXL-PCR and subsequent sequence analysis also resolved genotype discord in a mother/daughter relationship by revealing the presence of the CYP2D6*59 allelic variant in both individuals. Finally, we demonstrated that the 2939G>A single-nucleotide polymorphism present on CYP2D6*59 interfered with the TaqMan genotype assay that detected 2850C>T, causing false genotype assignments. Assay interference was resolved using an alternative TaqMan genotype assay currently available as a custom-made assay. These examples demonstrate the utility of ASXL-PCR for improved CYP2D6 allele/haplotype characterization. This fast, easy-to-perform method is not limited to CYP2D6 but can be adapted to any gene locus for which polymorphic sites are known.
Copyright © 2015 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Zhou S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin Pharmacokinet. 2009;48:689–723. - PubMed
-
- Zhou S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48:761–804. - PubMed
-
- Sim S.C., Ingelman-Sundberg M. Update on Allele Nomenclature for Human Cytochromes P450 and the Human Cytochrome P450 Allele (CYP-Allele) Nomenclature Database. Methods Mol Biol. 2013;987:251–259. - PubMed
-
- Teh L.K., Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27:55–67. - PubMed
-
- Zanger U.M., Raimundo S., Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

