Errors in RNA-Seq quantification affect genes of relevance to human disease
- PMID: 26335491
- PMCID: PMC4558956
- DOI: 10.1186/s13059-015-0734-x
Errors in RNA-Seq quantification affect genes of relevance to human disease
Abstract
Background: RNA-Seq has emerged as the standard for measuring gene expression and is an important technique often used in studies of human disease. Gene expression quantification involves comparison of the sequenced reads to a known genomic or transcriptomic reference. The accuracy of that quantification relies on there being enough unique information in the reads to enable bioinformatics tools to accurately assign the reads to the correct gene.
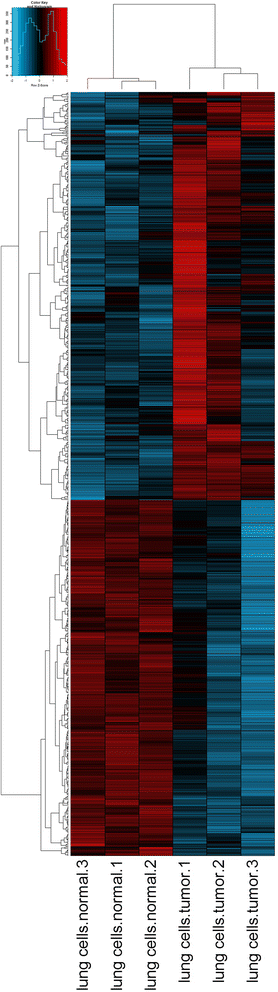
Results: We apply 12 common methods to estimate gene expression from RNA-Seq data and show that there are hundreds of genes whose expression is underestimated by one or more of those methods. Many of these genes have been implicated in human disease, and we describe their roles. We go on to propose a two-stage analysis of RNA-Seq data in which multi-mapped or ambiguous reads can instead be uniquely assigned to groups of genes. We apply this method to a recently published mouse cancer study, and demonstrate that we can extract relevant biological signal from data that would otherwise have been discarded.
Conclusions: For hundreds of genes in the human genome, RNA-Seq is unable to measure expression accurately. These genes are enriched for gene families, and many of them have been implicated in human disease. We show that it is possible to use data that may otherwise have been discarded to measure group-level expression, and that such data contains biologically relevant information.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004243/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004235/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900740/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

