Genome mining for natural product biosynthetic gene clusters in the Subsection V cyanobacteria
- PMID: 26335778
- PMCID: PMC4558948
- DOI: 10.1186/s12864-015-1855-z
Genome mining for natural product biosynthetic gene clusters in the Subsection V cyanobacteria
Abstract
Background: Cyanobacteria are well known for the production of a range of secondary metabolites. Whilst recent genome sequencing projects has led to an increase in the number of publically available cyanobacterial genomes, the secondary metabolite potential of many of these organisms remains elusive. Our study focused on the 11 publically available Subsection V cyanobacterial genomes, together with the draft genomes of Westiella intricata UH strain HT-29-1 and Hapalosiphon welwitschii UH strain IC-52-3, for their genetic potential to produce secondary metabolites. The Subsection V cyanobacterial genomes analysed in this study are reported to produce a diverse range of natural products, including the hapalindole-family of compounds, microcystin, hapalosin, mycosporine-like amino acids and hydrocarbons.
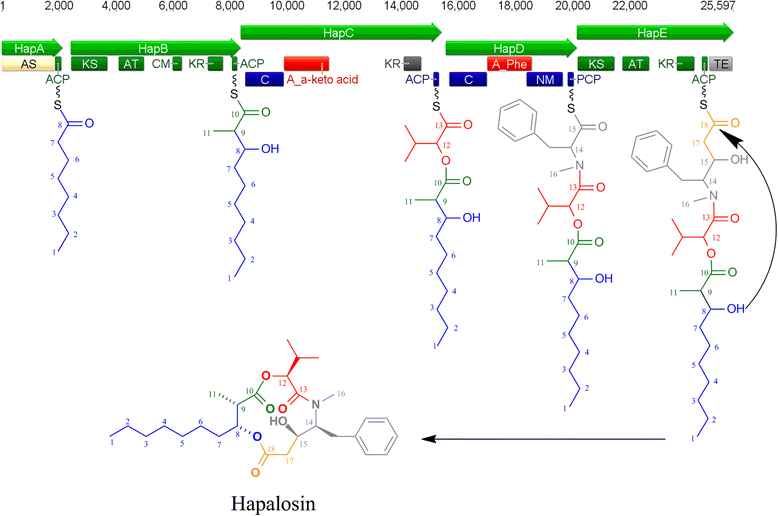
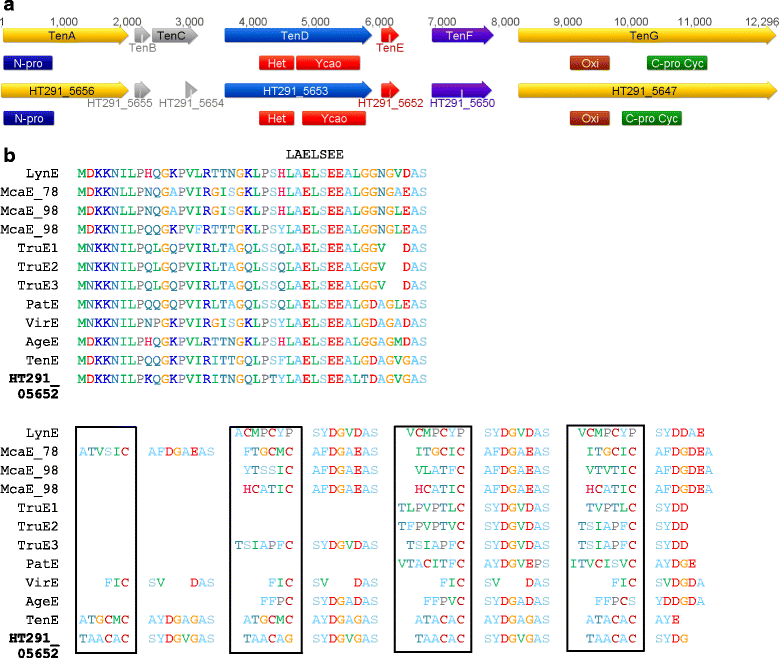
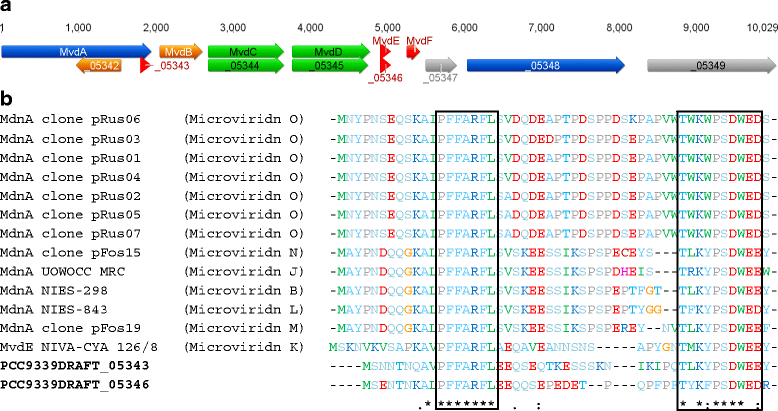
Results: A putative gene cluster for the cyclic depsipeptide hapalosin, known to reverse P-glycoprotein multiple drug resistance, was identified within three Subsection V cyanobacterial genomes, including the producing cyanobacterium H. welwitschii UH strain IC-52-3. A number of orphan NRPS/PKS gene clusters and ribosomally-synthesised and post translationally-modified peptide gene clusters (including cyanobactin, microviridin and bacteriocin gene clusters) were identified. Furthermore, gene clusters encoding the biosynthesis of mycosporine-like amino acids, scytonemin, hydrocarbons and terpenes were also identified and compared.
Conclusions: Genome mining has revealed the diversity, abundance and complex nature of the secondary metabolite potential of the Subsection V cyanobacteria. This bioinformatic study has identified novel biosynthetic enzymes which have not been associated with gene clusters of known classes of natural products, suggesting that these cyanobacteria potentially produce structurally novel secondary metabolites.
Figures



Similar articles
-
Comparative analysis of hapalindole, ambiguine and welwitindolinone gene clusters and reconstitution of indole-isonitrile biosynthesis from cyanobacteria.BMC Microbiol. 2014 Aug 1;14:213. doi: 10.1186/s12866-014-0213-7. BMC Microbiol. 2014. PMID: 25198896 Free PMC article.
-
Exploring cyanobacterial genomes for natural product biosynthesis pathways.Mar Genomics. 2015 Jun;21:1-12. doi: 10.1016/j.margen.2014.11.009. Epub 2014 Dec 5. Mar Genomics. 2015. PMID: 25482899 Review.
-
A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain.BMC Genomics. 2009 Aug 25;10:396. doi: 10.1186/1471-2164-10-396. BMC Genomics. 2009. PMID: 19706155 Free PMC article.
-
Genome-guided investigation of secondary metabolites produced by a potential new strain Streptomyces BA2 isolated from an endemic plant rhizosphere in Turkey.Arch Microbiol. 2021 Jul;203(5):2431-2438. doi: 10.1007/s00203-021-02210-z. Epub 2021 Mar 5. Arch Microbiol. 2021. PMID: 33666690
-
Unique marine derived cyanobacterial biosynthetic genes for chemical diversity.Nat Prod Rep. 2016 Feb;33(2):348-64. doi: 10.1039/c5np00097a. Nat Prod Rep. 2016. PMID: 26758451 Review.
Cited by
-
Actinobacteria From Desert: Diversity and Biotechnological Applications.Front Microbiol. 2021 Dec 9;12:765531. doi: 10.3389/fmicb.2021.765531. eCollection 2021. Front Microbiol. 2021. PMID: 34956128 Free PMC article. Review.
-
Recovering Genomics Clusters of Secondary Metabolites from Lakes Using Genome-Resolved Metagenomics.Front Microbiol. 2018 Feb 20;9:251. doi: 10.3389/fmicb.2018.00251. eCollection 2018. Front Microbiol. 2018. PMID: 29515540 Free PMC article.
-
Four new suomilides isolated from the cyanobacterium Nostoc sp. KVJ20 and proposal of their biosynthetic origin.Front Microbiol. 2023 Apr 20;14:1130018. doi: 10.3389/fmicb.2023.1130018. eCollection 2023. Front Microbiol. 2023. PMID: 37152725 Free PMC article.
-
Tandem ketone reduction in pepstatin biosynthesis reveals an F420H2-dependent statine pathway.Nat Commun. 2025 May 15;16(1):4531. doi: 10.1038/s41467-025-59785-0. Nat Commun. 2025. PMID: 40374670 Free PMC article.
-
Efficient Production of Shinorine and Mycosporine-Glycine-Alanine in Yarrowia lipolytica Using Natural and Engineered Nonribosomal Peptide Synthetases.J Agric Food Chem. 2025 Jul 2;73(26):16479-16489. doi: 10.1021/acs.jafc.5c03130. Epub 2025 Jun 21. J Agric Food Chem. 2025. PMID: 40542473 Free PMC article.
References
-
- Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P, Gould SB, Goremykin VV, Rippka R, Tandeau de Marsac N, et al. Genomes of Stigonematalean cyanobacteria (Subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 2013;5(1):31–44. doi: 10.1093/gbe/evs117. - DOI - PMC - PubMed
-
- Namikoshi M, Rinehart KL. Bioactive compounds produced by cyanobacteria. J Ind Microbiol Biotechnol. 1996;17(5–6):373–384. doi: 10.1007/BF01574768. - DOI
-
- Burja AM, Banaigs B, Abou-Mansour E, Grant Burgess J, Wright PC. Marine cyanobacteria—a prolific source of natural products. Tetrahedron. 2001;57(46):9347–9377. doi: 10.1016/S0040-4020(01)00931-0. - DOI
-
- Bhat V, Dave A, MacKay JA, Rawal VH: Chapter Two - The chemistry of hapalindoles, fischerindoles, ambiguines, and welwitindolinones. In: The Alkaloids: Chemistry and Biology. Edited by Hans-Joachim K, vol. Volume 73: New York, Academic Press; 2014: 65–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

