NCBI BLAST+ integrated into Galaxy
- PMID: 26336600
- PMCID: PMC4557756
- DOI: 10.1186/s13742-015-0080-7
NCBI BLAST+ integrated into Galaxy
Abstract
Background: The NCBI BLAST suite has become ubiquitous in modern molecular biology and is used for small tasks such as checking capillary sequencing results of single PCR products, genome annotation or even larger scale pan-genome analyses. For early adopters of the Galaxy web-based biomedical data analysis platform, integrating BLAST into Galaxy was a natural step for sequence comparison workflows.
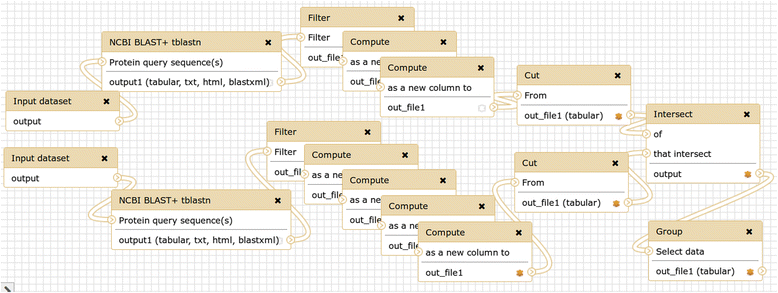
Findings: The command line NCBI BLAST+ tool suite was wrapped for use within Galaxy. Appropriate datatypes were defined as needed. The integration of the BLAST+ tool suite into Galaxy has the goal of making common BLAST tasks easy and advanced tasks possible.
Conclusions: This project is an informal international collaborative effort, and is deployed and used on Galaxy servers worldwide. Several examples of applications are described here.
Keywords: Accessibility; Annotation; BLAST; Galaxy; Pipeline; Reproducibility; Sequence analysis; Workflow.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

