Data integration in biological research: an overview
- PMID: 26336651
- PMCID: PMC4557916
- DOI: 10.1186/s40709-015-0032-5
Data integration in biological research: an overview
Abstract
Data sharing, integration and annotation are essential to ensure the reproducibility of the analysis and interpretation of the experimental findings. Often these activities are perceived as a role that bioinformaticians and computer scientists have to take with no or little input from the experimental biologist. On the contrary, biological researchers, being the producers and often the end users of such data, have a big role in enabling biological data integration. The quality and usefulness of data integration depend on the existence and adoption of standards, shared formats, and mechanisms that are suitable for biological researchers to submit and annotate the data, so it can be easily searchable, conveniently linked and consequently used for further biological analysis and discovery. Here, we provide background on what is data integration from a computational science point of view, how it has been applied to biological research, which key aspects contributed to its success and future directions.
Keywords: Bioinformatics; Data driven; Data integration; Open sciences; Standards.
Figures
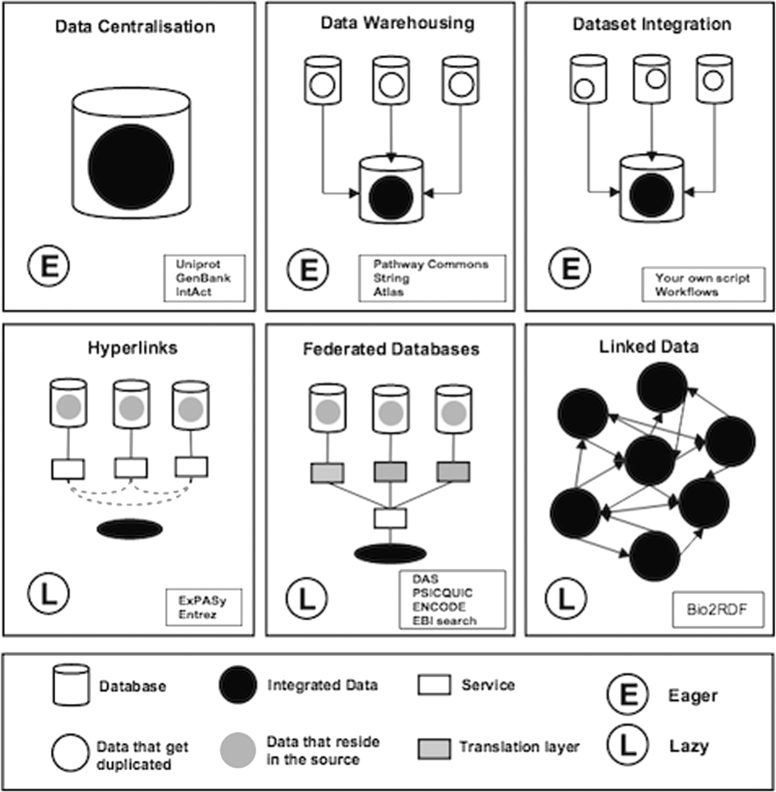
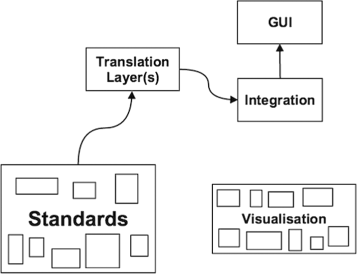
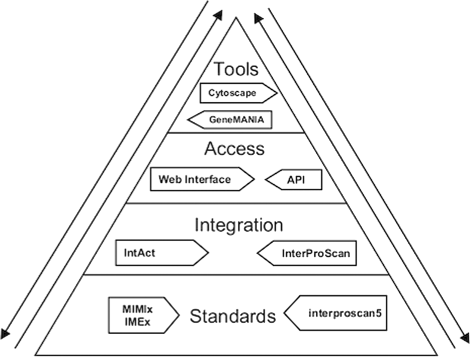



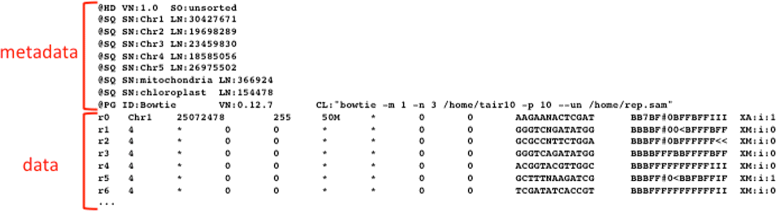
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

