Regulation of BDNF chromatin status and promoter accessibility in a neural correlate of associative learning
- PMID: 26336984
- PMCID: PMC5055205
- DOI: 10.1080/15592294.2015.1090072
Regulation of BDNF chromatin status and promoter accessibility in a neural correlate of associative learning
Abstract
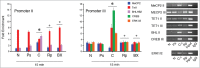
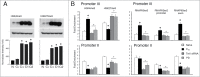
Brain-derived neurotrophic factor (BDNF) gene expression critically controls learning and its aberrant regulation is implicated in Alzheimer's disease and a host of neurodevelopmental disorders. The BDNF gene is target of known DNA regulatory mechanisms but details of its activity-dependent regulation are not fully characterized. We performed a comprehensive analysis of the epigenetic regulation of the turtle BDNF gene (tBDNF) during a neural correlate of associative learning using an in vitro model of eye blink classical conditioning. Shortly after conditioning onset, the results from ChIP-qPCR show conditioning-dependent increases in methyl-CpG-binding protein 2 (MeCP2) and repressor basic helix-loop-helix binding protein 2 (BHLHB2) binding to tBDNF promoter II that corresponds with transcriptional repression. In contrast, enhanced binding of ten-eleven translocation protein 1 (Tet1), extracellular signal-regulated kinase 1/2 (ERK1/2), and cAMP response element-binding protein (CREB) to promoter III corresponds with transcriptional activation. These actions are accompanied by rapid modifications in histone methylation and phosphorylation status of RNA polymerase II (RNAP II). Significantly, these remarkably coordinated changes in epigenetic factors for two alternatively regulated tBDNF promoters during conditioning are controlled by Tet1 and ERK1/2. Our findings indicate that Tet1 and ERK1/2 are critical partners that, through complementary functions, control learning-dependent tBDNF promoter accessibility required for rapid transcription and acquisition of classical conditioning.
Keywords: CREB; ChIP assays; Tet1; classical conditioning; histones.
Figures







References
-
- Peng S, Wuu J, Mufson EJ, Fahnestock M. Precursor form of BDNF and mature BDNF are decreased in the pre-clinical stages of Alzheiner's disease. J Neurochem 2005; 93:1412-21; PMID:15935057; http://dx.doi.org/10.1111/j.1471-4159.2005.03135.x - DOI - PubMed
-
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: When DNA methylation goes unrecognized. Nat Rev Genetics 2006; 7:415-26; PMID:16708070; http://dx.doi.org/10.1038/nrg1878 - DOI - PubMed
-
- Komulainen P, Pedersen M, Hanninin T, Bruunsgaard H, Lakka TA, Kivipelto M, Hassinen M, Rauramaa TH, Pedersen BK, Rauramaa R. BDNF is a novel marker of cognitive function in ageing women: The DR's EXTRA study. Neurobiol Learn Mem 2008; 90:596-603; http://dx.doi.org/10.1016/j.nlm.2008.07.014 - DOI - PubMed
-
- Guo JU, Su Y, Zhong C, Ming G, Song H. Hydroxylation of 5-methylcytosine by Tet1 promotes active DNA demethylation in the adult brain. Cell 2011; 145:423-34; PMID:21496894; http://dx.doi.org/10.1016/j.cell.2011.03.022 - DOI - PMC - PubMed
-
- Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14:341-56; PMID:23698584; http://dx.doi.org/10.1038/nrm3589 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
