A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins
- PMID: 26337144
- PMCID: PMC4757636
- DOI: 10.1002/anie.201505179
A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins
Abstract
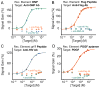
A versatile platform for the one-step fluorescence detection of both monovalent and multivalent proteins has been developed. This system is based on a conformation-switching stem-loop DNA scaffold that presents a small-molecule, polypeptide, or nucleic-acid recognition element on each of its two stem strands. The steric strain associated with the binding of one (multivalent) or two (monovalent) target molecules to these elements opens the stem, enhancing the emission of an attached fluorophore/quencher pair. The sensors respond rapidly (<10 min) and selectively, enabling the facile detection of specific proteins even in complex samples, such as blood serum. The versatility of the platform was demonstrated by detecting five bivalent proteins (four antibodies and the chemokine platelet-derived growth factor) and two monovalent proteins (a Fab fragment and the transcription factor TBP) with low nanomolar detection limits and no detectable cross-reactivity.
Keywords: DNA nanotechnology; antibodies; aptamers; fluorescence; molecular devices.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures




Similar articles
-
A Modular Nanoswitch for Mix-and-Detect Protein Assay Based on Binding-Induced Cascade Dissociation of Kissing Complex.Chembiochem. 2018 Apr 4;19(7):716-722. doi: 10.1002/cbic.201700649. Epub 2018 Feb 22. Chembiochem. 2018. PMID: 29356232
-
Investigation of molecular beacon aptamer-based bioassay for platelet-derived growth factor detection.Chembiochem. 2005 May;6(5):900-7. doi: 10.1002/cbic.200400308. Chembiochem. 2005. PMID: 15812865
-
Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay.Chembiochem. 2003 Sep 5;4(9):829-34. doi: 10.1002/cbic.200300615. Chembiochem. 2003. PMID: 12964156
-
Target-responsive structural switching for nucleic acid-based sensors.Acc Chem Res. 2010 May 18;43(5):631-41. doi: 10.1021/ar900245u. Acc Chem Res. 2010. PMID: 20222738 Review.
-
Molecular signaling aptamers for real-time fluorescence analysis of protein.IUBMB Life. 2005 Mar;57(3):123-8. doi: 10.1080/15216540500090686. IUBMB Life. 2005. PMID: 16036574 Review.
Cited by
-
Synthetic Antigen-Conjugated DNA Systems for Antibody Detection and Characterization.ACS Sens. 2023 Jul 28;8(7):2415-2426. doi: 10.1021/acssensors.3c00564. Epub 2023 Jul 18. ACS Sens. 2023. PMID: 37463359 Free PMC article. Review.
-
Nucleic Acids Analysis.Sci China Chem. 2021;64(2):171-203. doi: 10.1007/s11426-020-9864-7. Epub 2020 Dec 2. Sci China Chem. 2021. PMID: 33293939 Free PMC article. Review.
-
Programmable, Multiplexed DNA Circuits Supporting Clinically Relevant, Electrochemical Antibody Detection.ACS Sens. 2021 Jun 25;6(6):2442-2448. doi: 10.1021/acssensors.1c00790. Epub 2021 Jun 15. ACS Sens. 2021. PMID: 34129321 Free PMC article.
-
Antibody-powered nucleic acid release using a DNA-based nanomachine.Nat Commun. 2017 May 8;8:15150. doi: 10.1038/ncomms15150. Nat Commun. 2017. PMID: 28480878 Free PMC article.
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction.Nat Commun. 2023 Aug 14;14(1):4898. doi: 10.1038/s41467-023-40589-z. Nat Commun. 2023. PMID: 37580346 Free PMC article.

