Carbohydrate Structure Database: tools for statistical analysis of bacterial, plant and fungal glycomes
- PMID: 26337239
- PMCID: PMC4559136
- DOI: 10.1093/database/bav073
Carbohydrate Structure Database: tools for statistical analysis of bacterial, plant and fungal glycomes
Abstract
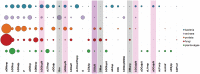
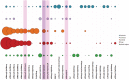
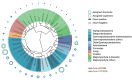
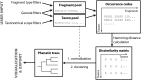
Carbohydrates are biological blocks participating in diverse and crucial processes both at cellular and organism levels. They protect individual cells, establish intracellular interactions, take part in the immune reaction and participate in many other processes. Glycosylation is considered as one of the most important modifications of proteins and other biologically active molecules. Still, the data on the enzymatic machinery involved in the carbohydrate synthesis and processing are scattered, and the advance on its study is hindered by the vast bulk of accumulated genetic information not supported by any experimental evidences for functions of proteins that are encoded by these genes. In this article, we present novel instruments for statistical analysis of glycomes in taxa. These tools may be helpful for investigating carbohydrate-related enzymatic activities in various groups of organisms and for comparison of their carbohydrate content. The instruments are developed on the Carbohydrate Structure Database (CSDB) platform and are available freely on the CSDB web-site at http://csdb.glycoscience.ru. Database URL: http://csdb.glycoscience.ru.
© The Author(s) 2015. Published by Oxford University Press.
Figures





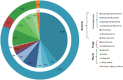

Similar articles
-
Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts.Nucleic Acids Res. 2016 Jan 4;44(D1):D1229-36. doi: 10.1093/nar/gkv840. Epub 2015 Aug 18. Nucleic Acids Res. 2016. PMID: 26286194 Free PMC article.
-
Bacterial, plant, and fungal carbohydrate structure databases: daily usage.Methods Mol Biol. 2015;1273:55-85. doi: 10.1007/978-1-4939-2343-4_5. Methods Mol Biol. 2015. PMID: 25753703
-
Expansion of coverage of Carbohydrate Structure Database (CSDB).Carbohydr Res. 2014 May 7;389:112-4. doi: 10.1016/j.carres.2013.10.009. Epub 2013 Oct 23. Carbohydr Res. 2014. PMID: 24680503
-
Enzymatic glycosylations on arrays.OMICS. 2010 Aug;14(4):437-44. doi: 10.1089/omi.2010.0035. OMICS. 2010. PMID: 20726800 Review.
-
Carbohydrate Structure Database: current state and recent developments.Anal Bioanal Chem. 2025 Feb;417(5):1025-1034. doi: 10.1007/s00216-024-05383-w. Epub 2024 Jun 25. Anal Bioanal Chem. 2025. PMID: 38914734 Review.
Cited by
-
Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts.Nucleic Acids Res. 2016 Jan 4;44(D1):D1229-36. doi: 10.1093/nar/gkv840. Epub 2015 Aug 18. Nucleic Acids Res. 2016. PMID: 26286194 Free PMC article.
-
In vivo manipulation of human gut Bacteroides fitness by abiotic oligosaccharides.Nat Chem Biol. 2025 Apr;21(4):544-554. doi: 10.1038/s41589-024-01763-6. Epub 2024 Oct 23. Nat Chem Biol. 2025. PMID: 39443715 Free PMC article.
-
Examining the diversity of structural motifs in fungal glycome.Comput Struct Biotechnol J. 2022 Sep 28;20:5466-5476. doi: 10.1016/j.csbj.2022.09.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36249563 Free PMC article.
-
Comparison of Methods for Bulk Automated Simulation of Glycosidic Bond Conformations.Int J Mol Sci. 2020 Oct 15;21(20):7626. doi: 10.3390/ijms21207626. Int J Mol Sci. 2020. PMID: 33076365 Free PMC article.
-
Source files of the Carbohydrate Structure Database: the way to sophisticated analysis of natural glycans.Sci Data. 2022 Mar 30;9(1):131. doi: 10.1038/s41597-022-01186-9. Sci Data. 2022. PMID: 35354826 Free PMC article.
References
-
- van Heijenoort J. (2001) Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology, 11, 25R–36R. - PubMed
-
- Latge J.P. (2007) The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol., 66, 279–290. - PubMed
-
- Selvendran R.R., O'Neill M.A. (1987) Isolation and Analysis of Cell Walls from Plant Material. John Wiley & Sons, Inc., Hoboken, NJ. - PubMed
-
- Baker J.L., Celik E., DeLisa M.P. (2013) Expanding the glycoengineering toolbox: the rise of bacterial N-linked protein glycosylation. Trends Biotechnol., 31, 313–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

