Piezoresistivity in single DNA molecules
- PMID: 26337293
- PMCID: PMC4569700
- DOI: 10.1038/ncomms9032
Piezoresistivity in single DNA molecules
Abstract
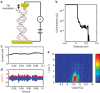
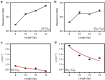
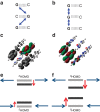
Piezoresistivity is a fundamental property of materials that has found many device applications. Here we report piezoresistivity in double helical DNA molecules. By studying the dependence of molecular conductance and piezoresistivity of single DNA molecules with different sequences and lengths, and performing molecular orbital calculations, we show that the piezoresistivity of DNA is caused by force-induced changes in the π-π electronic coupling between neighbouring bases, and in the activation energy of hole hopping. We describe the results in terms of thermal activated hopping model together with the ladder-based mechanical model for DNA proposed by de Gennes.
Figures


References
-
- Xu B., Xiao X. & Tao N. J. Measurements of single-molecule electromechanical properties. J. Am. Chem. Soc. 125, 16164–16165 (2003). - PubMed
-
- Diez-Perez I. et al. Controlling single-molecule conductance through lateral coupling of π orbitals. Nat. Nanotechnol. 6, 226–231 (2011). - PubMed
-
- Bruot C., Hihath J. & Tao N. Mechanically controlled molecular orbital alignment in single molecule junctions. Nat. Nanotechnol. 7, 35–40 (2012). - PubMed
-
- Rascón-Ramos H., Artés J. M., Li Y. & Hihath J. Binding configurations and intramolecular strain in single-molecule devices. Nat. Mater. 14, 1–6 (2015). - PubMed
-
- Rajski S. R., Jackson B. A. & Barton J. K. DNA repair: models for damage and mismatch recognition. Mutat. Res. 447, 49–72 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

