Proteasomal degradation of preemptive quality control (pQC) substrates is mediated by an AIRAPL-p97 complex
- PMID: 26337389
- PMCID: PMC4626058
- DOI: 10.1091/mbc.E15-02-0085
Proteasomal degradation of preemptive quality control (pQC) substrates is mediated by an AIRAPL-p97 complex
Abstract
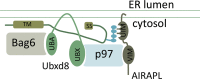
The initial folding of secreted proteins occurs in the ER lumen, which contains specific chaperones and where posttranslational modifications may occur. Therefore lack of translocation, regardless of entry route or protein identity, is a highly toxic event, as the newly synthesized polypeptide is misfolded and can promiscuously interact with cytosolic factors. Mislocalized proteins bearing a signal sequence that did not successfully translocate through the translocon complex are subjected to a preemptive quality control (pQC) pathway and are degraded by the ubiquitin-proteasome system (UPS). In contrast to UPS-mediated, ER-associated degradation, few components involved in pQC have been identified. Here we demonstrate that on specific translocation inhibition, a p97-AIRAPL complex directly binds and regulates the efficient processing of polyubiquitinated pQC substrates by the UPS. We also demonstrate p97's role in pQC processing of preproinsulin in cases of naturally occurring mutations within the signal sequence of insulin.
© 2015 Braunstein et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

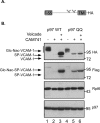
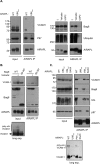
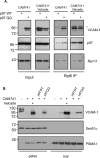
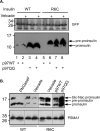
References
-
- Ast T, Schuldiner M. All roads lead to Rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit Rev Biochem Mol Biol. 2013;48:273–288. - PubMed
-
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, Whitney S, Board PG. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. - PubMed
-
- Besemer J, Harant H, Wang S, Oberhauser B, Marquardt K, Foster CA, Schreiner EP, de Vries JE, Dascher-Nadel C, Lindley IJ. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

