Ribosome profiling reveals an important role for translational control in circadian gene expression
- PMID: 26338483
- PMCID: PMC4665005
- DOI: 10.1101/gr.191296.115
Ribosome profiling reveals an important role for translational control in circadian gene expression
Abstract
Physiological and behavioral circadian rhythms are driven by a conserved transcriptional/translational negative feedback loop in mammals. Although most core clock factors are transcription factors, post-transcriptional control introduces delays that are critical for circadian oscillations. Little work has been done on circadian regulation of translation, so to address this deficit we conducted ribosome profiling experiments in a human cell model for an autonomous clock. We found that most rhythmic gene expression occurs with little delay between transcription and translation, suggesting that the lag in the accumulation of some clock proteins relative to their mRNAs does not arise from regulated translation. Nevertheless, we found that translation occurs in a circadian fashion for many genes, sometimes imposing an additional level of control on rhythmically expressed mRNAs and, in other cases, conferring rhythms on noncycling mRNAs. Most cyclically transcribed RNAs are translated at one of two major times in a 24-h day, while rhythmic translation of most noncyclic RNAs is phased to a single time of day. Unexpectedly, we found that the clock also regulates the formation of cytoplasmic processing (P) bodies, which control the fate of mRNAs, suggesting circadian coordination of mRNA metabolism and translation.
© 2015 Jang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
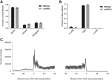
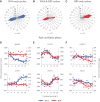
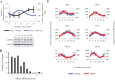

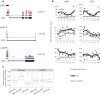
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
