The Genome of a Tortoise Herpesvirus (Testudinid Herpesvirus 3) Has a Novel Structure and Contains a Large Region That Is Not Required for Replication In Vitro or Virulence In Vivo
- PMID: 26339050
- PMCID: PMC4645648
- DOI: 10.1128/JVI.01794-15
The Genome of a Tortoise Herpesvirus (Testudinid Herpesvirus 3) Has a Novel Structure and Contains a Large Region That Is Not Required for Replication In Vitro or Virulence In Vivo
Abstract
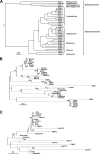
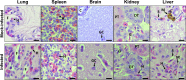
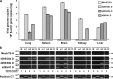
Testudinid herpesvirus 3 (TeHV-3) is the causative agent of a lethal disease affecting several tortoise species. The threat that this virus poses to endangered animals is focusing efforts on characterizing its properties, in order to enable the development of prophylactic methods. We have sequenced the genomes of the two most studied TeHV-3 strains (1976 and 4295). TeHV-3 strain 1976 has a novel genome structure and is most closely related to a turtle herpesvirus, thus supporting its classification into genus Scutavirus, subfamily Alphaherpesvirinae, family Herpesviridae. The sequence of strain 1976 also revealed viral counterparts of cellular interleukin-10 and semaphorin, which have not been described previously in members of subfamily Alphaherpesvirinae. TeHV-3 strain 4295 is a mixture of three forms (m1, m2, and M), in which, in comparison to strain 1976, the genomes exhibit large, partially overlapping deletions of 12.5 to 22.4 kb. Viral subclones representing these forms were isolated by limiting dilution assays, and each replicated in cell culture comparably to strain 1976. With the goal of testing the potential of the three forms as attenuated vaccine candidates, strain 4295 was inoculated intranasally into Hermann's tortoises (Testudo hermanni). All inoculated subjects died, and PCR analyses demonstrated the ability of the m2 and M forms to spread and invade the brain. In contrast, the m1 form was detected in none of the organs tested, suggesting its potential as the basis of an attenuated vaccine candidate. Our findings represent a major step toward characterizing TeHV-3 and developing prophylactic methods against it.
Importance: Testudinid herpesvirus 3 (TeHV-3) causes a lethal disease in tortoises, several species of which are endangered. We have characterized the viral genome and used this information to take steps toward developing an attenuated vaccine. We have sequenced the genomes of two strains (1976 and 4295), compared their growth in vitro, and investigated the pathogenesis of strain 4295, which consists of three deletion mutants. The major findings are that (i) TeHV-3 has a novel genome structure, (ii) its closest relative is a turtle herpesvirus, (iii) it contains interleukin-10 and semaphorin genes (the first time these have been reported in an alphaherpesvirus), (iv) a sizeable region of the genome is not required for viral replication in vitro or virulence in vivo, and (v) one of the components of strain 4295, which has a deletion of 22.4 kb, exhibits properties indicating that it may serve as the starting point for an attenuated vaccine.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures







References
-
- Pellett PE, Davison AJ, Eberle R, Ehlers B, Hayward GS, Lacoste V, Minson AC, Nicholas J, Roizman B, Studdert MJ, Wang F. 2012. Order—Herpesvirales, p 99–107. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Elsevier, San Diego, CA. doi:10.1016/B978-0-12-384684-6.00005-7. - DOI
-
- Pellett PE, Roizman B. 2013. Herpesviridae, p 1802–1822. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Origgi FC. 2006. Herpesvirus in tortoises, p 814–821. In Mader DR. (ed), Reptile medicine and surgery, 2nd ed Saunders, St. Louis, MO.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

