Molecular docking based screening of neem-derived compounds with the NS1 protein of Influenza virus
- PMID: 26339153
- PMCID: PMC4546996
- DOI: 10.6026/97320630011359
Molecular docking based screening of neem-derived compounds with the NS1 protein of Influenza virus
Abstract
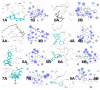
Different strains of influenza virus are affecting a large number of people worldwide to combat with Influenza virus destruction, numerous synthetic antiviral medicines are available for influenza virus in the market. But still there was a need for the development of drug which will target all the strains of influenza virus. For this purpose conserved residues within the influenza virus NS1 protein have been found by aligning all the available sequences of existing strains from the national center of biotechnology information(NCBI) protein database. The compounds from leaf extracts of neem (Azadirachta indica), previously known to have antiviral properties, were virtually screened to identify side effects free natural drug. Molecular docking identified eight potential compounds (Tetratriacontane, 127-40-2, 6-o-ACETYLNIMBANDIOL, Rutin, Tiplasinin, Hyperoside, ( )- Nimocinolide and Quercitrin) found to have perfect binding with reported conserved residues (R19, R35, S42 and D39) of influenza virus NS1 protein involved in the binding of drugs. From, further analysis 6-o-ACETYLNIMBANDIOL, Rutin and Tiplasinin were found as drug against influenza strains because their binding residues were conserved in all strains. The potential of neem chemical against influenza virus has best been highlighted through this study and it provides direction for further consideration of these products for in-vivo and in-vitro validations.
Abbreviations: NS1 protein - Non Structural 1 protein, NA - Neuraminidase, HA - Hemagglutinin, M - Yersinia enterocolitica 8081, ypk - Yersinia pestis KIM, yps - Yersinia pseudotuberculosis Db1, KEGG - Kyoto Encyclopedia of Genes and Genomes, KO - KEGG Orthology, KOBAS - KO Based Annotation System.
Keywords: Influenza virus; Molecular docking; NS1 protein; Neem leaf extract.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
