A host factor supports retrotransposition of the TRE5-A population in Dictyostelium cells by suppressing an Argonaute protein
- PMID: 26339297
- PMCID: PMC4559204
- DOI: 10.1186/s13100-015-0045-5
A host factor supports retrotransposition of the TRE5-A population in Dictyostelium cells by suppressing an Argonaute protein
Abstract
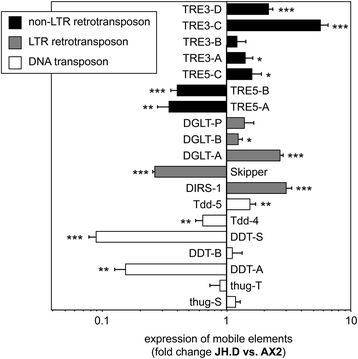
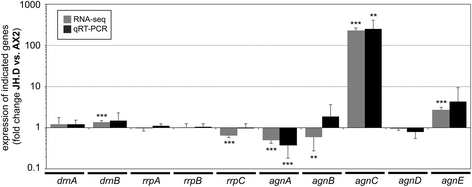
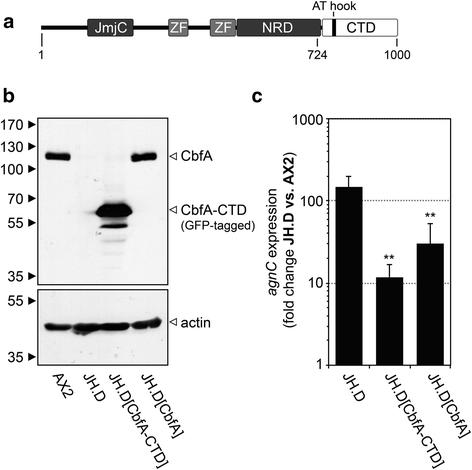
Background: In the compact and haploid genome of Dictyostelium discoideum control of transposon activity is of particular importance to maintain viability. The non-long terminal repeat retrotransposon TRE5-A amplifies continuously in D. discoideum cells even though it produces considerable amounts of minus-strand (antisense) RNA in the presence of an active RNA interference machinery. Removal of the host-encoded C-module-binding factor (CbfA) from D. discoideum cells resulted in a more than 90 % reduction of both plus- and minus-strand RNA of TRE5-A and a strong decrease of the retrotransposition activity of the cellular TRE5-A population. Transcriptome analysis revealed an approximately 230-fold overexpression of the gene coding for the Argonaute-like protein AgnC in a CbfA-depleted mutant.
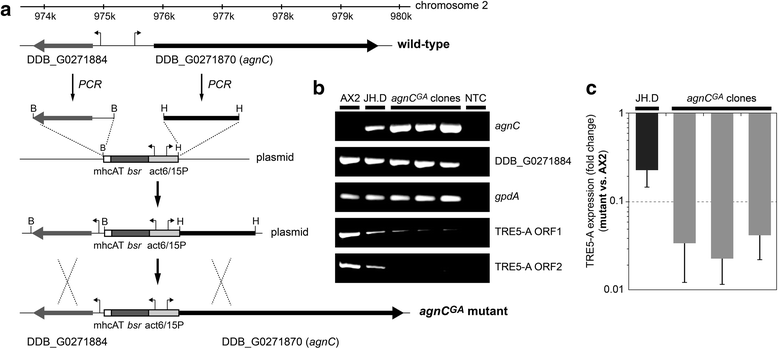
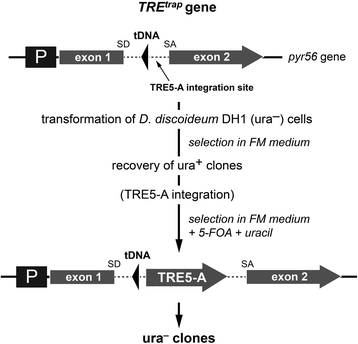
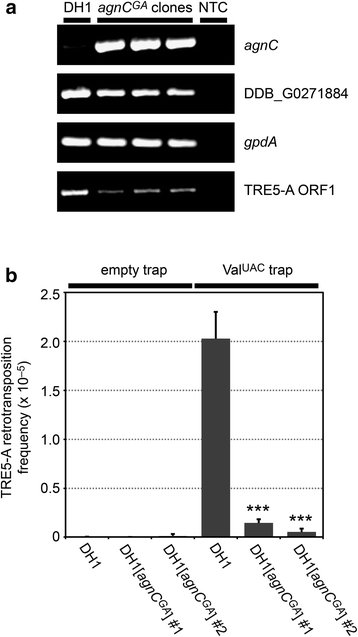
Results: The D. discoideum genome contains orthologs of RNA-dependent RNA polymerases, Dicer-like proteins, and Argonaute proteins that are supposed to represent RNA interference pathways. We analyzed available mutants in these genes for altered expression of TRE5-A. We found that the retrotransposon was overexpressed in mutants lacking the Argonaute proteins AgnC and AgnE. Because the agnC gene is barely expressed in wild-type cells, probably due to repression by CbfA, we employed a new method of promoter-swapping to overexpress agnC in a CbfA-independent manner. In these strains we established an in vivo retrotransposition assay that determines the retrotransposition frequency of the cellular TRE5-A population. We observed that both the TRE5-A steady-state RNA level and retrotransposition rate dropped to less than 10 % of wild-type in the agnC overexpressor strains.
Conclusions: The data suggest that TRE5-A amplification is controlled by a distinct pathway of the Dictyostelium RNA interference machinery that does not require RNA-dependent RNA polymerases but involves AgnC. This control is at least partially overcome by the activity of CbfA, a factor derived from the retrotransposon's host. This unusual regulation of mobile element activity most likely had a profound effect on genome evolution in D. discoideum.
Keywords: Argonaute; Dictyostelium; RNAi; Retrotransposition; siRNA.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

