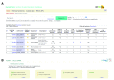
SwissPalm: Protein Palmitoylation database
- PMID: 26339475
- PMCID: PMC4544385
- DOI: 10.12688/f1000research.6464.1
SwissPalm: Protein Palmitoylation database
Abstract
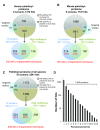
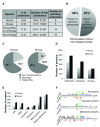
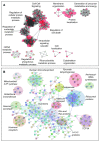
Protein S-palmitoylation is a reversible post-translational modification that regulates many key biological processes, although the full extent and functions of protein S-palmitoylation remain largely unexplored. Recent developments of new chemical methods have allowed the establishment of palmitoyl-proteomes of a variety of cell lines and tissues from different species. As the amount of information generated by these high-throughput studies is increasing, the field requires centralization and comparison of this information. Here we present SwissPalm ( http://swisspalm.epfl.ch), our open, comprehensive, manually curated resource to study protein S-palmitoylation. It currently encompasses more than 5000 S-palmitoylated protein hits from seven species, and contains more than 500 specific sites of S-palmitoylation. SwissPalm also provides curated information and filters that increase the confidence in true positive hits, and integrates predictions of S-palmitoylated cysteine scores, orthologs and isoform multiple alignments. Systems analysis of the palmitoyl-proteome screens indicate that 10% or more of the human proteome is susceptible to S-palmitoylation. Moreover, ontology and pathway analyses of the human palmitoyl-proteome reveal that key biological functions involve this reversible lipid modification. Comparative analysis finally shows a strong crosstalk between S-palmitoylation and other post-translational modifications. Through the compilation of data and continuous updates, SwissPalm will provide a powerful tool to unravel the global importance of protein S-palmitoylation.
Keywords: Acyl-RAC; Acyl-biotin exchange; S-palmitoylation; database; palmitoyl-proteomes; proteomics.
Conflict of interest statement
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

