Dynamic Water-Mediated Hydrogen Bonding in a Collagen Model Peptide
- PMID: 26339765
- PMCID: PMC4648280
- DOI: 10.1021/acs.biochem.5b00622
Dynamic Water-Mediated Hydrogen Bonding in a Collagen Model Peptide
Abstract
In the canonical (G-X-Y)(n) sequence of the fibrillar collagen triple helix, stabilizing direct interchain hydrogen bonding connects neighboring chains. Mutations of G can disrupt these interactions and are linked to connective tissue diseases. Here we integrate computational approaches with nuclear magnetic resonance (NMR) to obtain a dynamic view of hydrogen bonding distributions in the (POG)(4)(-)(POA)-(POG)(5) peptide, showing that the solution conformation, dynamics, and hydrogen bonding deviate from the reported X-ray crystal structure in many aspects. The simulations and NMR data provide clear evidence of inequivalent environments in the three chains. Molecular dynamics (MD) simulations indicate direct interchain hydrogen bonds in the leading chain, water bridges in the middle chain, and nonbridging waters in the trailing chain at the G → A substitution site. Theoretical calculations of NMR chemical shifts using a quantum fragmentation procedure can account for the unusual downfield NMR chemical shifts at the substitution sites and are used to assign the resonances to the individual chains. The NMR and MD data highlight the sensitivity of amide shifts to changes in the acceptor group from peptide carbonyls to water. The results are used to interpret solution NMR data for a variety of glycine substitutions and other sequence triplet interruptions to provide new connections between collagen sequences, their associated structures, dynamical behavior, and their ability to recognize collagen receptors.
Figures
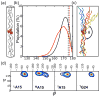
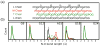


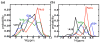

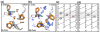
References
-
- Myllyharju J, Kivirikko KI. Collagens, Modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. - PubMed
-
- Heino J. The collagen family members as cell adhesion proteins. Bioessays. 2007;29:1001–1010. - PubMed
-
- Ramachandran GN, Kartha G. Structure of collagen. Nature. 1955;176:593–595. - PubMed
-
- Rich A, Crick FH. The structure of collagen. Nature. 1955;176:915–916. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

