Mercury Reduction and Methyl Mercury Degradation by the Soil Bacterium Xanthobacter autotrophicus Py2
- PMID: 26341208
- PMCID: PMC4616949
- DOI: 10.1128/AEM.01982-15
Mercury Reduction and Methyl Mercury Degradation by the Soil Bacterium Xanthobacter autotrophicus Py2
Abstract
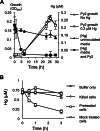
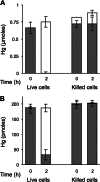
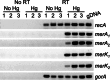
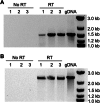
Two previously uncharacterized potential broad-spectrum mercury (Hg) resistance operons (mer) are present on the chromosome of the soil Alphaproteobacteria Xanthobacter autotrophicus Py2. These operons, mer1 and mer2, contain two features which are commonly found in mer operons in the genomes of soil and marine Alphaproteobacteria, but are not present in previously characterized mer operons: a gene for the mercuric reductase (MerA) that encodes an alkylmercury lyase domain typical of those found on the MerB protein, and the presence of an additional gene, which we are calling merK, with homology to glutathione reductase. Here, we demonstrate that Py2 is resistant to 0.2 μM inorganic mercury [Hg(II)] and 0.05 μM methylmercury (MeHg). Py2 is capable of converting MeHg and Hg(II) to elemental mercury [Hg(0)], and reduction of Hg(II) is induced by incubation in sub toxic concentrations of Hg(II). Transcription of the merA genes increased with Hg(II) treatment, and in both operons merK resides on the same polycistronic mRNA as merA. We propose the use of Py2 as a model system for studying the contribution of mer to Hg mobility in soil and marine ecosystems.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Organomercurial Lyase (MerB)-Mediated Demethylation Decreases Bacterial Methylmercury Resistance in the Absence of Mercuric Reductase (MerA).Appl Environ Microbiol. 2022 Mar 22;88(6):e0001022. doi: 10.1128/aem.00010-22. Epub 2022 Feb 9. Appl Environ Microbiol. 2022. PMID: 35138926 Free PMC article.
-
Anaerobic Mercury Methylation and Demethylation by Geobacter bemidjiensis Bem.Environ Sci Technol. 2016 Apr 19;50(8):4366-73. doi: 10.1021/acs.est.6b00401. Epub 2016 Apr 7. Environ Sci Technol. 2016. PMID: 27019098
-
The local impact of a coal-fired power plant on inorganic mercury and methyl-mercury distribution in rice (Oryza sativa L.).Environ Pollut. 2017 Apr;223:11-18. doi: 10.1016/j.envpol.2016.11.042. Epub 2017 Jan 28. Environ Pollut. 2017. PMID: 28139322
-
Bacterial resistances to inorganic mercury salts and organomercurials.Plasmid. 1992 Jan;27(1):4-16. doi: 10.1016/0147-619x(92)90002-r. Plasmid. 1992. PMID: 1311113 Review.
-
Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts.FASEB J. 1988 Feb;2(2):124-30. doi: 10.1096/fasebj.2.2.3277886. FASEB J. 1988. PMID: 3277886 Review.
Cited by
-
Genetic and Physiological Adaptations of Marine Bacterium Pseudomonas stutzeri 273 to Mercury Stress.Front Microbiol. 2018 Apr 5;9:682. doi: 10.3389/fmicb.2018.00682. eCollection 2018. Front Microbiol. 2018. PMID: 29675016 Free PMC article.
-
Microbial Contributions to Heavy Metal Phytoremediation in Agricultural Soils: A Review.Microorganisms. 2024 Sep 25;12(10):1945. doi: 10.3390/microorganisms12101945. Microorganisms. 2024. PMID: 39458255 Free PMC article. Review.
-
Glutathione reductase plays a role in the metabolism of methylmercury degradation in Rhodotorula mucilaginosa.Microbiol Spectr. 2025 Feb 4;13(2):e0239524. doi: 10.1128/spectrum.02395-24. Epub 2025 Jan 16. Microbiol Spectr. 2025. PMID: 39817752 Free PMC article.
-
Biotransformation of As, Cr, Hg, and Mn by Pseudomonadota: chances and risks.Biodegradation. 2025 Jul 15;36(4):60. doi: 10.1007/s10532-025-10157-x. Biodegradation. 2025. PMID: 40663258 Free PMC article. Review.
-
Mercury in Soil and Forage Plants from Artisanal and Small-Scale Gold Mining in the Bombana Area, Indonesia.Toxics. 2020 Feb 22;8(1):15. doi: 10.3390/toxics8010015. Toxics. 2020. PMID: 32098420 Free PMC article.
References
-
- Ullrich SM, Tanton TW, Abdrashitova SA. 2001. Mercury in the aquatic environment: a review of factors affecting methylation. Crit Rev Environ Sci Technol 31:241–293. doi:10.1080/20016491089226. - DOI
-
- Roman HA, Walsh TL, Coull BA, Dewailly È Guallar E, Hattis D, Mariën K, Schwartz J, Stern AH, Virtanen JK, Rice G. 2011. Evaluation of the cardiovascular effects of methylmercury exposures: current evidence supports development of a dose-response function for regulatory benefits analysis. Environ Health Perspect 119:607–614. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

