A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear
- PMID: 26341477
- PMCID: PMC4775449
- DOI: 10.1016/j.heares.2015.08.013
A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear
Abstract
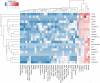
The mammalian inner ear consists of the cochlea and the vestibular labyrinth (utricle, saccule, and semicircular canals), which participate in both hearing and balance. Proper development and life-long function of these structures involves a highly complex coordinated system of spatial and temporal gene expression. The characterization of the inner ear transcriptome is likely important for the functional study of auditory and vestibular components, yet, primarily due to tissue unavailability, detailed expression catalogues of the human inner ear remain largely incomplete. We report here, for the first time, comprehensive transcriptome characterization of the adult human cochlea, ampulla, saccule and utricle of the vestibule obtained from patients without hearing abnormalities. Using RNA-Seq, we measured the expression of >50,000 predicted genes corresponding to approximately 200,000 transcripts, in the adult inner ear and compared it to 32 other human tissues. First, we identified genes preferentially expressed in the inner ear, and unique either to the vestibule or cochlea. Next, we examined expression levels of specific groups of potentially interesting RNAs, such as genes implicated in hearing loss, long non-coding RNAs, pseudogenes and transcripts subject to nonsense mediated decay (NMD). We uncover the spatial specificity of expression of these RNAs in the hearing/balance system, and reveal evidence of tissue specific NMD. Lastly, we investigated the non-syndromic deafness loci to which no gene has been mapped, and narrow the list of potential candidates for each locus. These data represent the first high-resolution transcriptome catalogue of the adult human inner ear. A comprehensive identification of coding and non-coding RNAs in the inner ear will enable pathways of auditory and vestibular function to be further defined in the study of hearing and balance. Expression data are freely accessible at https://www.tgen.org/home/research/research-divisions/neurogenomics/supplementary-data/inner-ear-transcriptome.aspx.
Keywords: Balance; Cochlea; Hearing; RNA-Seq; Transcriptome; Vestibule.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures




References
-
- Andrews S. FastQC a quality control tool for high throughput sequence data. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet. 1995;11:369–75. doi:10.1038/ng1295-369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

