Acquisition of Portal Venous Circulating Tumor Cells From Patients With Pancreaticobiliary Cancers by Endoscopic Ultrasound
- PMID: 26341722
- PMCID: PMC4985007
- DOI: 10.1053/j.gastro.2015.08.050
Acquisition of Portal Venous Circulating Tumor Cells From Patients With Pancreaticobiliary Cancers by Endoscopic Ultrasound
Abstract
Background & aims: Tumor cells circulate in low numbers in peripheral blood; their detection is used predominantly in metastatic disease. We evaluated the feasibility and safety of sampling portal venous blood via endoscopic ultrasound (EUS) to count portal venous circulating tumor cells (CTCs), compared with paired peripheral CTCs, in patients with pancreaticobiliary cancers (PBCs).
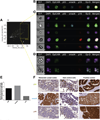
Methods: In a single-center cohort study, we evaluated 18 patients with suspected PBCs. Under EUS guidance, a 19-gauge EUS fine needle was advanced transhepatically into the portal vein and as many as four 7.5-mL aliquots of blood were aspirated. Paired peripheral blood samples were obtained. Epithelial-derived CTCs were sorted magnetically based on expression of epithelial cell adhesion molecules; only those with a proper morphology and found to be CD45 negative and positive for cytokeratins 8, 18, and/or 19 and 4',6-diamidino-2-phenylindole were considered to be CTCs. For 5 samples, CTCs also were isolated by flow cytometry and based on CD45 depletion. ImageStream was used to determine the relative protein levels of P16, SMAD4, and P53. DNA was extracted from CTCs for sequencing of select KRAS codons.
Results: There were no complications from portal vein blood acquisition. We detected CTCs in portal vein samples from all 18 patients (100%) vs peripheral blood samples from only 4 patients (22.2%). Patients with confirmed PBCs had a mean of 118.4 ± 36.8 CTCs/7.5 mL portal vein blood, compared with a mean of 0.8 ± 0.4 CTCs/7.5 mL peripheral blood (P < .01). The 9 patients with nonmetastatic, resectable, or borderline-resectable PBCs had a mean of 83.2 CTCs/7.5 mL portal vein blood (median, 62.0 CTCs/7.5 mL portal vein blood). In a selected patient, portal vein CTCs were found to carry the same mutations as those detected in a metastatic lymph node and expressed similar levels of P16, SMAD4, and P53 proteins.
Conclusions: It is feasible and safe to collect portal venous blood from patients undergoing EUS. We identified CTCs in all portal vein blood samples from patients with PBCs, but less than 25% of peripheral blood samples. Portal vein CTCs can be used for molecular characterization of PBCs and share features of metastatic tissue. This technique might be used to study the pathogenesis and progression of PBCs, as well as a diagnostic or prognostic tool to stratify risk of cancer recurrence or developing metastases.
Keywords: Endoscopic Ultrasound; EpCAM; Pancreatic Cancer; Portal Venous Blood.
Copyright © 2015 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors disclose no conflicts.
Figures



References
-
- Howlader NNA, Krapcho M, Garshell J, et al. SEER Cancer Statistics Review, 1975–2012, National Cancer Institute. Bethesda, MD: [Accessed: June 10, 2015]. Available from: http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission.
-
- Neoptolemos JP, Stocken DD, Friess H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–1210. - PubMed
-
- Oettle H, Post S, Neuhaus P, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. - PubMed
-
- Barugola G, Falconi M, Bettini R, et al. The determinant factors of recurrence following resection for ductal pancreatic cancer. JOP. 2007;8:132–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

