The Mediator complex subunits MED25/PFT1 and MED8 are required for transcriptional responses to changes in cell wall arabinose composition and glucose treatment in Arabidopsis thaliana
- PMID: 26341899
- PMCID: PMC4560864
- DOI: 10.1186/s12870-015-0592-4
The Mediator complex subunits MED25/PFT1 and MED8 are required for transcriptional responses to changes in cell wall arabinose composition and glucose treatment in Arabidopsis thaliana
Abstract
Background: Plant cell walls are dynamic structures involved in all aspects of plant growth, environmental interactions and defense responses, and are the most abundant renewable source of carbon-containing polymers on the planet. To balance rigidity and extensibility, the composition and integrity of cell wall components need to be tightly regulated, for example during cell elongation.
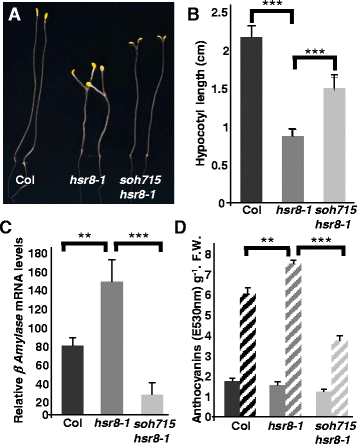
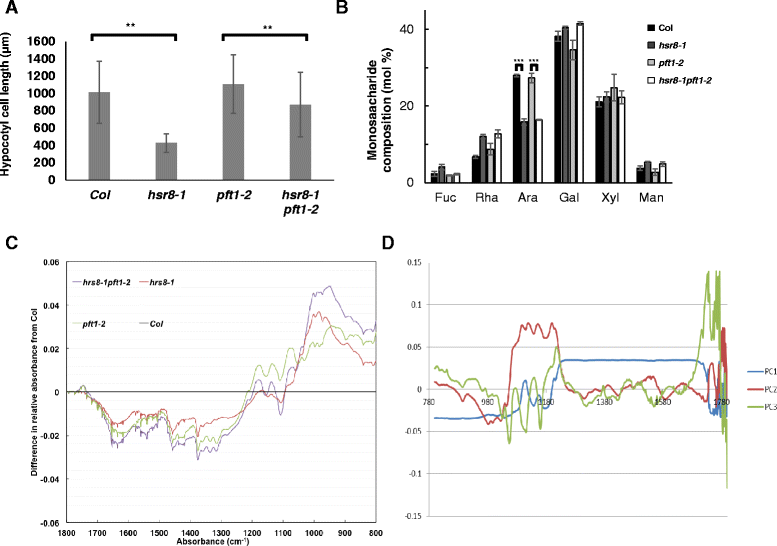
Results: We show that mutations in the MED25/PFT1 and MED8 subunits of the Mediator transcription complex suppressed the sugar-hypersensitive hypocotyl elongation phenotype of the hsr8-1 mutant, which has cell wall defects due to arabinose deficiency that do not permit normal cell elongation. This suppression occurred independently of light and jasmonic acid (JA) signaling. Gene expression analyses revealed that the expression of genes induced in hsr8-1 that encode enzymes and proteins that are involved in cell expansion and cell wall strengthening is reduced in the pft1-2 mutant line, and the expression of genes encoding transcription factors involved in reducing hypocotyl cell elongation, genes encoding cell wall associated enzymes and proteins is up-regulated in pft1-2. PFT1 was also required for the expression of several glucose-induced genes, including those encoding cell wall components and enzymes, regulatory and enzymatic components of anthocyanin biosynthesis, and flavonoid and glucosinolate biosynthetic pathways.
Conclusions: These results establish that MED25 and MED8 subunits of the Mediator transcriptional complex are required for the transcriptional regulation of genes involved in cell elongation and cell wall composition in response to defective cell walls and in sugar- responsive gene expression.
Figures




References
-
- Reiter W-D. Biosynthesis and properties of the plant cell wall. Curr Opin Plant Biol. 2002;5:536–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/000CA386/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E01772X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/1356X/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F007582/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I001271/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004588/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E022758/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G17764/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G18881/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

