Birds Generally Carry a Small Repertoire of Bitter Taste Receptor Genes
- PMID: 26342138
- PMCID: PMC4607536
- DOI: 10.1093/gbe/evv180
Birds Generally Carry a Small Repertoire of Bitter Taste Receptor Genes
Abstract
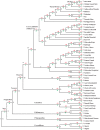
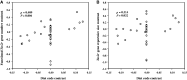
As they belong to the most species-rich class of tetrapod vertebrates, birds have long been believed to possess an inferior taste system. However, the bitter taste is fundamental in birds to recognize dietary toxins (which are typically bitter) in potential food sources. To characterize the evolution of avian bitter taste receptor genes (Tas2rs) and to test whether dietary toxins have shaped the repertoire size of avian Tas2rs, we examined 48 genomes representing all but 3 avian orders. The total number of Tas2r genes was found to range from 1 in the domestic pigeon to 12 in the bar-tailed trogon, with an average of 4, which suggested that a much smaller Tas2r gene repertoire exists in birds than in other vertebrates. Furthermore, we uncovered a positive correlation between the number of putatively functional Tas2rs and the abundance of potential toxins in avian diets. Because plant products contain more toxins than animal tissues and insects release poisonous defensive secretions, we hypothesized that herbivorous and insectivorous birds may demand more functional Tas2rs than carnivorous birds feeding on noninsect animals. Our analyses appear to support this hypothesis and highlight the critical role of taste perception in birds.
Keywords: Tas2r; birds; bitter taste; diet; feeding ecology.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes.Mol Biol Evol. 2014 Dec;31(12):3216-27. doi: 10.1093/molbev/msu254. Epub 2014 Aug 31. Mol Biol Evol. 2014. PMID: 25180257
-
Characterization and phylogeny of bitter taste receptor genes (Tas2r) in Squamata.Genetica. 2019 Apr;147(2):131-139. doi: 10.1007/s10709-019-00056-4. Epub 2019 Mar 6. Genetica. 2019. PMID: 30840187
-
Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire.Mol Biol Evol. 2014 Feb;31(2):303-9. doi: 10.1093/molbev/mst219. Epub 2013 Nov 7. Mol Biol Evol. 2014. PMID: 24202612 Free PMC article.
-
The repertoire of bitter taste receptor genes in canids.Amino Acids. 2017 Jul;49(7):1159-1167. doi: 10.1007/s00726-017-2422-5. Epub 2017 Apr 17. Amino Acids. 2017. PMID: 28417226 Review.
-
Research progress of the bitter taste receptor genes in primates.Yi Chuan. 2018 Feb 20;40(2):126-134. doi: 10.16288/j.yczz.17-258. Yi Chuan. 2018. PMID: 29428905 Review.
Cited by
-
The evolution of bitter taste receptor gene in primates: Gene duplication and selection.Ecol Evol. 2023 Oct 13;13(10):e10610. doi: 10.1002/ece3.10610. eCollection 2023 Oct. Ecol Evol. 2023. PMID: 37841228 Free PMC article.
-
Comparative and population genomics approaches reveal the basis of adaptation to deserts in a small rodent.Mol Ecol. 2020 Apr;29(7):1300-1314. doi: 10.1111/mec.15401. Epub 2020 Mar 23. Mol Ecol. 2020. PMID: 32130752 Free PMC article.
-
Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats.Mol Ecol. 2018 Nov;27(22):4475-4488. doi: 10.1111/mec.14873. Epub 2018 Nov 3. Mol Ecol. 2018. PMID: 30230081 Free PMC article.
-
Activation of the Nucleus Taeniae of the Amygdala by Umami Taste in Domestic Chicks (Gallus gallus).Front Physiol. 2022 May 26;13:897931. doi: 10.3389/fphys.2022.897931. eCollection 2022. Front Physiol. 2022. PMID: 35694389 Free PMC article.
-
Transcriptome sequencing and phylogenetic analysis of four species of luminescent beetles.Sci Rep. 2017 May 12;7(1):1814. doi: 10.1038/s41598-017-01835-9. Sci Rep. 2017. PMID: 28500331 Free PMC article.
References
-
- Adler E, et al. 2000. A novel family of mammalian taste receptors. Cell 100:693–702. - PubMed
-
- Behrens M, Korsching SI, Meyerhof W. 2014. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol Biol Evol. 31:3216–3227. - PubMed
-
- Berkhoudt H. 1985. Structure and function of avian taste recetors. In: Levy AS, McLelland J, editors. Form and function in birds III. New York: Academic press; p. 463–496.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

