Desiderata for computable representations of electronic health records-driven phenotype algorithms
- PMID: 26342218
- PMCID: PMC4639716
- DOI: 10.1093/jamia/ocv112
Desiderata for computable representations of electronic health records-driven phenotype algorithms
Abstract
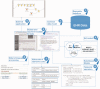
Background: Electronic health records (EHRs) are increasingly used for clinical and translational research through the creation of phenotype algorithms. Currently, phenotype algorithms are most commonly represented as noncomputable descriptive documents and knowledge artifacts that detail the protocols for querying diagnoses, symptoms, procedures, medications, and/or text-driven medical concepts, and are primarily meant for human comprehension. We present desiderata for developing a computable phenotype representation model (PheRM).
Methods: A team of clinicians and informaticians reviewed common features for multisite phenotype algorithms published in PheKB.org and existing phenotype representation platforms. We also evaluated well-known diagnostic criteria and clinical decision-making guidelines to encompass a broader category of algorithms.
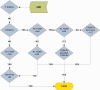
Results: We propose 10 desired characteristics for a flexible, computable PheRM: (1) structure clinical data into queryable forms; (2) recommend use of a common data model, but also support customization for the variability and availability of EHR data among sites; (3) support both human-readable and computable representations of phenotype algorithms; (4) implement set operations and relational algebra for modeling phenotype algorithms; (5) represent phenotype criteria with structured rules; (6) support defining temporal relations between events; (7) use standardized terminologies and ontologies, and facilitate reuse of value sets; (8) define representations for text searching and natural language processing; (9) provide interfaces for external software algorithms; and (10) maintain backward compatibility.
Conclusion: A computable PheRM is needed for true phenotype portability and reliability across different EHR products and healthcare systems. These desiderata are a guide to inform the establishment and evolution of EHR phenotype algorithm authoring platforms and languages.
Keywords: computable representation; data models; electronic health records; phenotype algorithms; phenotype standardization.
© The Author 2015. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures
References
-
- Li L, Ruau D, Chen R, et al. Systematic identification of risk factors for Alzheimer’s disease through shared genetic architecture and electronic medical records. Pac Symp Biocomput Pac Symp Biocomput. 2013;2013:224–235. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HG006385/HG/NHGRI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01-HG006388/HG/NHGRI NIH HHS/United States
- U01-HG006389/HG/NHGRI NIH HHS/United States
- U01-HG006385/HG/NHGRI NIH HHS/United States
- U01 HG004603/HG/NHGRI NIH HHS/United States
- U01 HG004610/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG004608/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01-HG004610/HG/NHGRI NIH HHS/United States
- R01 GM103859/GM/NIGMS NIH HHS/United States
- U01-HG006375/HG/NHGRI NIH HHS/United States
- U01-HG04603/HG/NHGRI NIH HHS/United States
- U01-HG004608/HG/NHGRI NIH HHS/United States
- R01 LM010685/LM/NLM NIH HHS/United States
- R01-LM010685/LM/NLM NIH HHS/United States
- U54 HD083211/HD/NICHD NIH HHS/United States
- U01 HG004609/HG/NHGRI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- U01-HG04599/HG/NHGRI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- U01 HG004599/HG/NHGRI NIH HHS/United States
- U01-HG06379/HG/NHGRI NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- U01-HG006378/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- U01-HG004609/HG/NHGRI NIH HHS/United States
- R01 GM105688/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

