DENdb: database of integrated human enhancers
- PMID: 26342387
- PMCID: PMC4560934
- DOI: 10.1093/database/bav085
DENdb: database of integrated human enhancers
Abstract
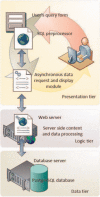
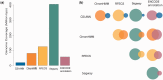
Enhancers are cis-acting DNA regulatory regions that play a key role in distal control of transcriptional activities. Identification of enhancers, coupled with a comprehensive functional analysis of their properties, could improve our understanding of complex gene transcription mechanisms and gene regulation processes in general. We developed DENdb, a centralized on-line repository of predicted enhancers derived from multiple human cell-lines. DENdb integrates enhancers predicted by five different methods generating an enriched catalogue of putative enhancers for each of the analysed cell-lines. DENdb provides information about the overlap of enhancers with DNase I hypersensitive regions, ChIP-seq regions of a number of transcription factors and transcription factor binding motifs, means to explore enhancer interactions with DNA using several chromatin interaction assays and enhancer neighbouring genes. DENdb is designed as a relational database that facilitates fast and efficient searching, browsing and visualization of information. Database URL: http://www.cbrc.kaust.edu.sa/dendb/.
© The Author(s) 2015. Published by Oxford University Press.
Figures

References
-
- Lee T.I., Young R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. - PubMed
-
- Maston G.A., Evans S.K., Green M.R. (2006) Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet., 7, 29–59. - PubMed
-
- Shlyueva D., Stampfel G., Stark A. (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet., 15, 272–286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

