Co-option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty
- PMID: 26343453
- PMCID: PMC4573913
- DOI: 10.1016/j.devcel.2015.08.005
Co-option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty
Abstract
The evolutionary origins of complex morphological structures such as the vertebrate eye or insect wing remain one of the greatest mysteries of biology. Recent comparative studies of gene expression imply that new structures are not built from scratch, but rather form by co-opting preexisting gene networks. A key prediction of this model is that upstream factors within the network will activate their preexisting targets (i.e., enhancers) to form novel anatomies. Here, we show how a recently derived morphological novelty present in the genitalia of D. melanogaster employs an ancestral Hox-regulated network deployed in the embryo to generate the larval posterior spiracle. We demonstrate how transcriptional enhancers and constituent transcription factor binding sites are used in both ancestral and novel contexts. These results illustrate network co-option at the level of individual connections between regulatory genes and highlight how morphological novelty may originate through the co-option of networks controlling seemingly unrelated structures.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
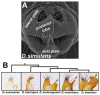


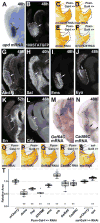


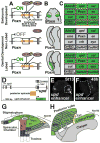
Comment in
-
Evolving Genital Structures: A Deep Look at Network Co-option.Dev Cell. 2015 Sep 14;34(5):485-6. doi: 10.1016/j.devcel.2015.08.022. Dev Cell. 2015. PMID: 26374760
References
-
- Abouheif E. Establishing homology criteria for regulatory gene networks: prospects and challenges. Novartis Found Symp. 1999;222:207–221. discussion 222–225. - PubMed
-
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. - PubMed
-
- Barrio R, Shea MJ, Carulli J, Lipkow K, Gaul U, Frommer G, Schuh R, Jäckle H, Kafatos FC. The spalt-related gene of Drosophila melanogaster is a member of an ancient gene family, defined by the adjacent, region-specific homeotic gene spalt. Dev Genes Evol. 1996;206:315–325. - PubMed
-
- Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

