Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA
- PMID: 26343537
- PMCID: PMC4669199
- DOI: 10.1038/ni.3267
Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA
Abstract
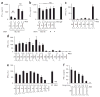
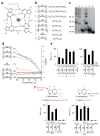
Cytosolic DNA that emerges during infection with a retrovirus or DNA virus triggers antiviral type I interferon responses. So far, only double-stranded DNA (dsDNA) over 40 base pairs (bp) in length has been considered immunostimulatory. Here we found that unpaired DNA nucleotides flanking short base-paired DNA stretches, as in stem-loop structures of single-stranded DNA (ssDNA) derived from human immunodeficiency virus type 1 (HIV-1), activated the type I interferon-inducing DNA sensor cGAS in a sequence-dependent manner. DNA structures containing unpaired guanosines flanking short (12- to 20-bp) dsDNA (Y-form DNA) were highly stimulatory and specifically enhanced the enzymatic activity of cGAS. Furthermore, we found that primary HIV-1 reverse transcripts represented the predominant viral cytosolic DNA species during early infection of macrophages and that these ssDNAs were highly immunostimulatory. Collectively, our study identifies unpaired guanosines in Y-form DNA as a highly active, minimal cGAS recognition motif that enables detection of HIV-1 ssDNA.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



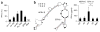
Comment in
-
Reading the fine print: sequence-specific activation of cGAS.Nat Immunol. 2015 Oct;16(10):1009-10. doi: 10.1038/ni.3274. Nat Immunol. 2015. PMID: 26382862 No abstract available.
References
-
- Krieg AM, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. - PubMed
-
- Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. - PubMed
-
- Hartmann G, et al. Rational design of new CpG oligonucleotides that combine B cell activation with high IFN-alpha induction in plasmacytoid dendritic cells. Eur J Immunol. 2003;33:1633–1641. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

