ATM functions at the peroxisome to induce pexophagy in response to ROS
- PMID: 26344566
- PMCID: PMC4589490
- DOI: 10.1038/ncb3230
ATM functions at the peroxisome to induce pexophagy in response to ROS
Abstract
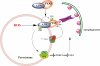
Peroxisomes are highly metabolic, autonomously replicating organelles that generate reactive oxygen species (ROS) as a by-product of fatty acid β-oxidation. Consequently, cells must maintain peroxisome homeostasis, or risk pathologies associated with too few peroxisomes, such as peroxisome biogenesis disorders, or too many peroxisomes, inducing oxidative damage and promoting diseases such as cancer. We report that the PEX5 peroxisome import receptor binds ataxia-telangiectasia mutated (ATM) and localizes this kinase to the peroxisome. In response to ROS, ATM signalling activates ULK1 and inhibits mTORC1 to induce autophagy. Specificity for autophagy of peroxisomes (pexophagy) is provided by ATM phosphorylation of PEX5 at Ser 141, which promotes PEX5 monoubiquitylation at Lys 209, and recognition of ubiquitylated PEX5 by the autophagy adaptor protein p62, directing the autophagosome to peroxisomes to induce pexophagy. These data reveal an important new role for ATM in metabolism as a sensor of ROS that regulates pexophagy.
Figures
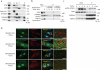
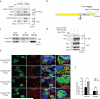



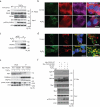
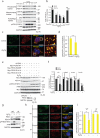
Comment in
-
A mammalian pexophagy target.Nat Cell Biol. 2015 Nov;17(11):1371-3. doi: 10.1038/ncb3253. Epub 2015 Oct 12. Nat Cell Biol. 2015. PMID: 26458245 Free PMC article.
Similar articles
-
A new role for ATM in selective autophagy of peroxisomes (pexophagy).Autophagy. 2016;12(4):711-2. doi: 10.1080/15548627.2015.1123375. Autophagy. 2016. PMID: 27050462 Free PMC article.
-
PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS.Autophagy. 2023 Jun;19(6):1781-1802. doi: 10.1080/15548627.2022.2160566. Epub 2023 Jan 1. Autophagy. 2023. PMID: 36541703 Free PMC article.
-
A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS.Nat Cell Biol. 2013 Oct;15(10):1186-96. doi: 10.1038/ncb2822. Epub 2013 Aug 18. Nat Cell Biol. 2013. PMID: 23955302 Free PMC article.
-
Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis.Cell Cycle. 2017;16(21):2037-2045. doi: 10.1080/15384101.2017.1376149. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933989 Free PMC article. Review.
-
Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders.Autophagy. 2017 May 4;13(5):991-994. doi: 10.1080/15548627.2017.1291480. Epub 2017 Feb 28. Autophagy. 2017. PMID: 28318378 Free PMC article. Review.
Cited by
-
PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation.J Cell Biol. 2016 Sep 12;214(6):677-90. doi: 10.1083/jcb.201511034. Epub 2016 Sep 5. J Cell Biol. 2016. PMID: 27597759 Free PMC article.
-
Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy.J Cell Biol. 2019 Mar 4;218(3):798-807. doi: 10.1083/jcb.201804172. Epub 2019 Jan 30. J Cell Biol. 2019. PMID: 30700497 Free PMC article.
-
Mitophagy in the Pathogenesis of Liver Diseases.Cells. 2020 Mar 30;9(4):831. doi: 10.3390/cells9040831. Cells. 2020. PMID: 32235615 Free PMC article. Review.
-
Reciprocity in ROS and autophagic signaling.Curr Opin Toxicol. 2018 Feb;7:28-36. doi: 10.1016/j.cotox.2017.10.006. Epub 2017 Oct 12. Curr Opin Toxicol. 2018. PMID: 29457143 Free PMC article.
-
mTORC1/2 inhibitor and curcumin induce apoptosis through lysosomal membrane permeabilization-mediated autophagy.Oncogene. 2018 Sep;37(38):5205-5220. doi: 10.1038/s41388-018-0345-6. Epub 2018 May 30. Oncogene. 2018. PMID: 29849119 Free PMC article.
References
-
- Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annual review of nutrition. 2001;21:193–230. - PubMed
-
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. - PubMed
-
- Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. - PubMed
-
- Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature reviews. Molecular cell biology. 2014;15:411–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

