Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration
- PMID: 26344735
- PMCID: PMC4769684
- DOI: 10.1016/j.preteyeres.2015.09.002
Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration
Abstract
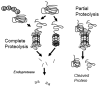
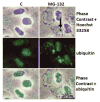
Maintenance of protein homeostasis, also referred to as "Proteostasis", integrates multiple pathways that regulate protein synthesis, folding, translocation, and degradation. Failure in proteostasis may be one of the underlying mechanisms responsible for the cascade of events leading to age-related macular degeneration (AMD). This review covers the major degradative pathways (ubiquitin-proteasome and lysosomal involvement in phagocytosis and autophagy) in the retinal pigment epithelium (RPE) and summarizes evidence of their involvement in AMD. Degradation of damaged and misfolded proteins via the proteasome occurs in coordination with heat shock proteins. Evidence of increased content of proteasome and heat shock proteins in retinas from human donors with AMD is consistent with increased oxidative stress and extensive protein damage with AMD. Phagocytosis and autophagy share key molecules in phagosome maturation as well as degradation of their cargo following fusion with lysosomes. Phagocytosis and degradation of photoreceptor outer segments ensures functional integrity of the neural retina. Autophagy rids the cell of toxic protein aggregates and defective mitochondria. Evidence suggesting a decline in autophagic flux includes the accumulation of autophagic substrates and damaged mitochondria in RPE from AMD donors. An age-related decrease in lysosomal enzymatic activity inhibits autophagic clearance of outer segments, mitochondria, and protein aggregates, thereby accelerating the accumulation of lipofuscin. This cumulative damage over a person's lifetime tips the balance in RPE from a state of para-inflammation, which strives to restore cell homeostasis, to the chronic inflammation associated with AMD.
Keywords: Age-related macular degeneration; Autophagy; Lysosome; Proteasome; Proteostasis; Retinal pigment epithelium.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
















References
-
- Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, Hauser MA, Rickman CB, Bok D, Hageman GS, Johnson LV. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. - PMC - PubMed
-
- Agarwal AK, Xing C, DeMartino GN, Mizrachi D, Hernandez MD, Sousa AB, Martinez de Villarreal L, dos Santos HH, Garg A. PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am J Hum Genet. 2010;87:866–872. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

