A quantitative description of Ndc80 complex linkage to human kinetochores
- PMID: 26345214
- PMCID: PMC4569735
- DOI: 10.1038/ncomms9161
A quantitative description of Ndc80 complex linkage to human kinetochores
Abstract
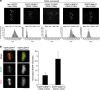
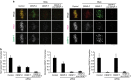
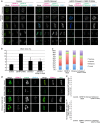
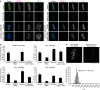
The Ndc80 complex, which mediates end-on attachment of spindle microtubules, is linked to centromeric chromatin in human cells by two inner kinetochore proteins, CENP-T and CENP-C. Here to quantify their relative contributions to Ndc80 recruitment, we combine measurements of kinetochore protein copy number with selective protein depletion assays. This approach reveals about 244 Ndc80 complexes per human kinetochore (∼14 per kinetochore microtubule), 215 CENP-C, 72 CENP-T and only 151 Ndc80s as part of the KMN protein network (1:1:1 Knl1, Mis12 and Ndc80 complexes). Each CENP-T molecule recruits ∼2 Ndc80 complexes; one as part of a KMN network. In contrast, ∼40% of CENP-C recruits only a KMN network. Replacing the CENP-C domain that binds KMN with the CENP-T domain that recruits both an Ndc80 complex and KMN network yielded functional kinetochores. These results provide a quantitative picture of the linkages between centromeric chromatin and the microtubule-binding Ndc80 complex at the human kinetochore.
Figures


References
-
- Musacchio A. & Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 8, 379–393 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

