Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris
- PMID: 26346253
- PMCID: PMC4562351
- DOI: 10.1186/s12864-015-1863-z
Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris
Abstract
Background: Candida auris is a multidrug resistant, emerging agent of fungemia in humans. Its actual global distribution remains obscure as the current commercial methods of clinical diagnosis misidentify it as C. haemulonii. Here we report the first draft genome of C. auris to explore the genomic basis of virulence and unique differences that could be employed for differential diagnosis.
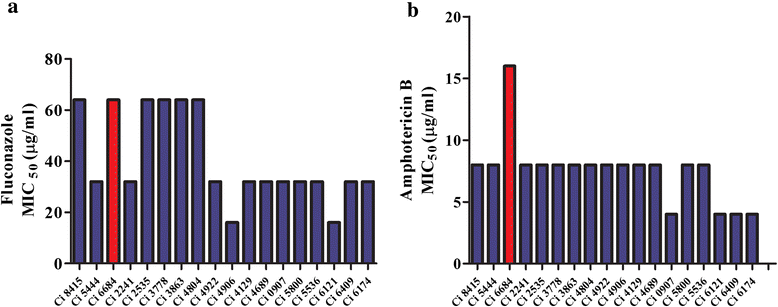
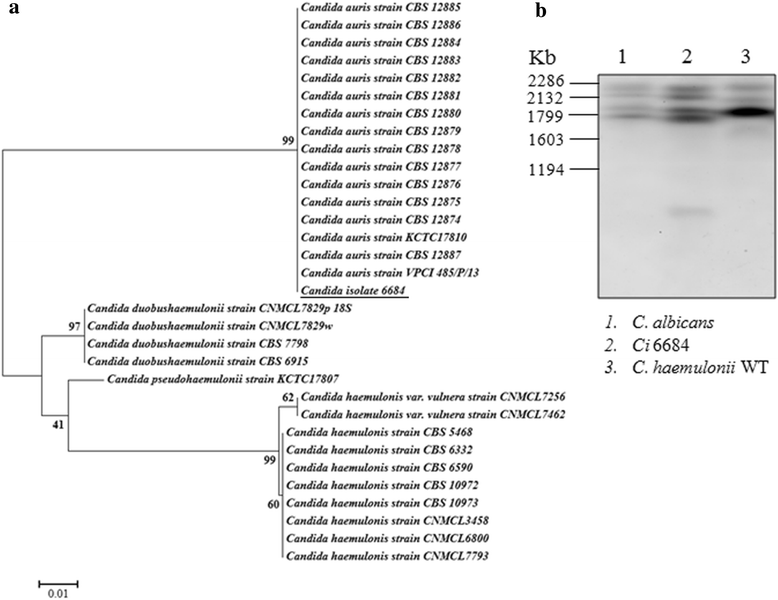
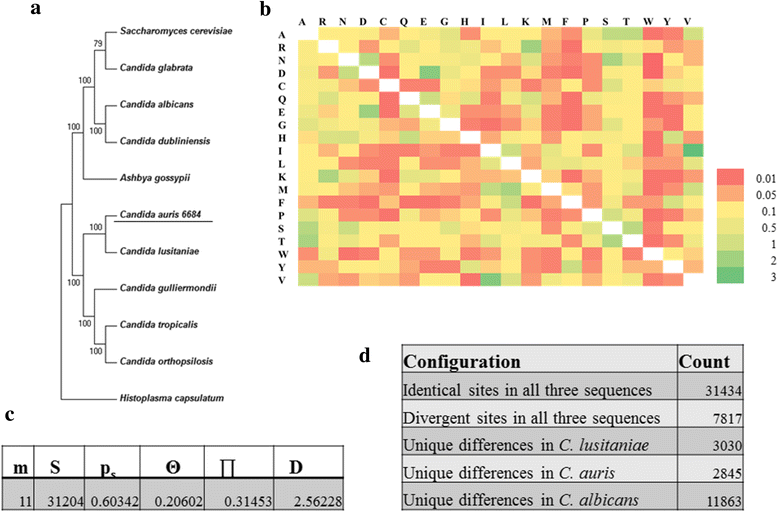
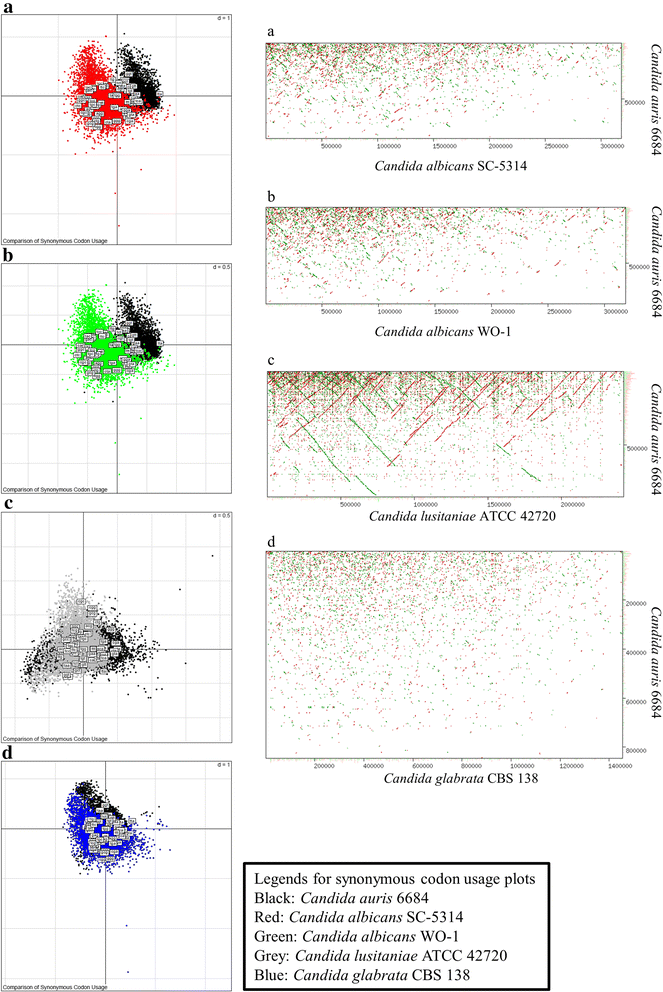
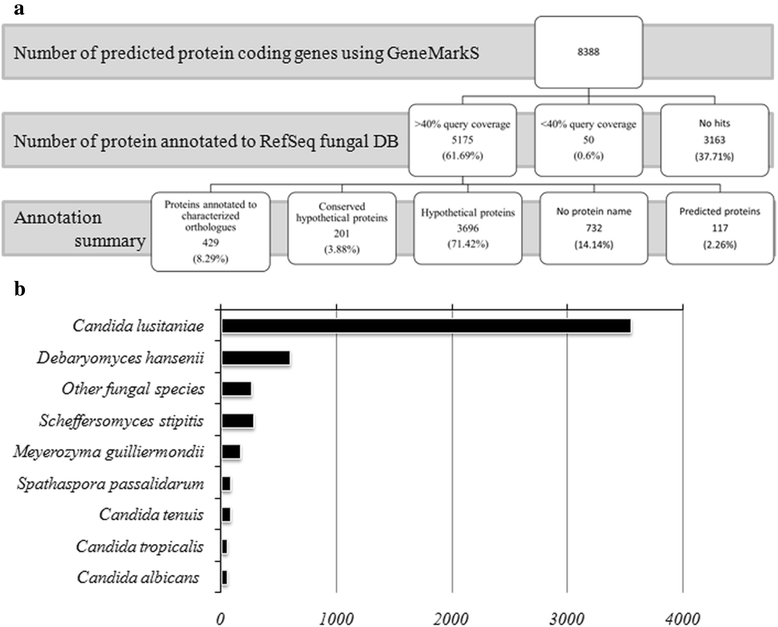
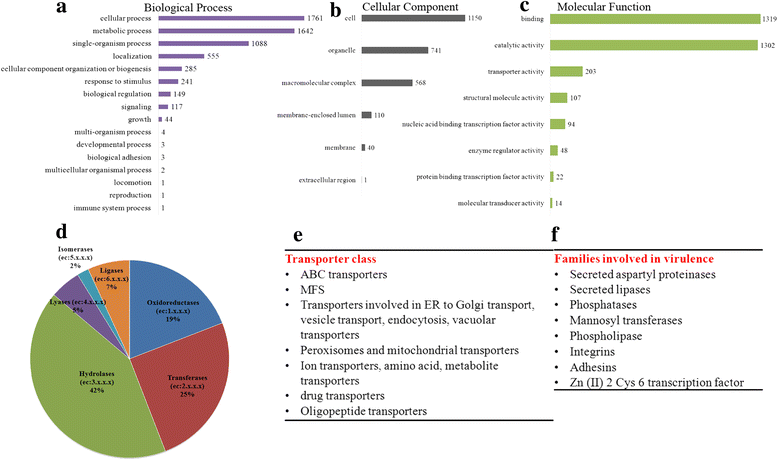
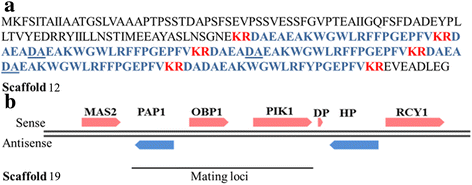
Results: More than 99.5 % of the C. auris genomic reads did not align to the current whole (or draft) genome sequences of Candida albicans, Candida lusitaniae, Candida glabrata and Saccharomyces cerevisiae; thereby indicating its divergence from the active Candida clade. The genome spans around 12.49 Mb with 8527 predicted genes. Functional annotation revealed that among the sequenced Candida species, it is closest to the hemiascomycete species Clavispora lusitaniae. Comparison with the well-studied species Candida albicans showed that it shares significant virulence attributes with other pathogenic Candida species such as oligopeptide transporters, mannosyl transfersases, secreted proteases and genes involved in biofilm formation. We also identified a plethora of transporters belonging to the ABC and major facilitator superfamily along with known MDR transcription factors which explained its high tolerance to antifungal drugs.
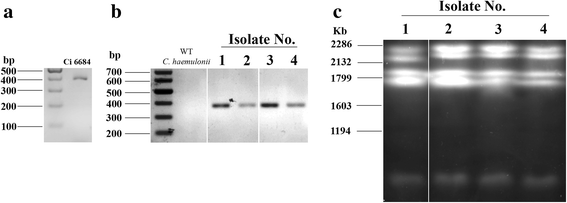
Conclusions: Our study emphasizes an urgent need for accurate fungal screening methods such as PCR and electrophoretic karyotyping to ensure proper management of fungemia. Our work highlights the potential genetic mechanisms involved in virulence and pathogenicity of an important emerging human pathogen namely C. auris. Owing to its diversity at the genomic scale; we expect the genome sequence to be a useful resource to map species specific differences that will help develop accurate diagnostic markers and better drug targets.
Figures
Similar articles
-
Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species.Nat Commun. 2018 Dec 17;9(1):5346. doi: 10.1038/s41467-018-07779-6. Nat Commun. 2018. PMID: 30559369 Free PMC article.
-
Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses.Clin Infect Dis. 2017 Jan 15;64(2):134-140. doi: 10.1093/cid/ciw691. Epub 2016 Oct 20. Clin Infect Dis. 2017. PMID: 27988485 Free PMC article.
-
Emergence of the novel sixth Candida auris Clade VI in Bangladesh.Microbiol Spectr. 2024 Jul 2;12(7):e0354023. doi: 10.1128/spectrum.03540-23. Epub 2024 Jun 6. Microbiol Spectr. 2024. PMID: 38842332 Free PMC article.
-
Candida auris: Epidemiology, biology, antifungal resistance, and virulence.PLoS Pathog. 2020 Oct 22;16(10):e1008921. doi: 10.1371/journal.ppat.1008921. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33091071 Free PMC article. Review.
-
Candida and candidaemia. Susceptibility and epidemiology.Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
Cited by
-
A Case of Multiple Myeloma Presenting as Streptococcus pneumoniae Meningitis with Candida auris Fungemia.Case Rep Oncol. 2018 Nov 1;11(3):705-710. doi: 10.1159/000493852. eCollection 2018 Sep-Dec. Case Rep Oncol. 2018. PMID: 30483102 Free PMC article.
-
Prevalence and Therapeutic Challenges of Fungal Drug Resistance: Role for Plants in Drug Discovery.Antibiotics (Basel). 2020 Mar 31;9(4):150. doi: 10.3390/antibiotics9040150. Antibiotics (Basel). 2020. PMID: 32244276 Free PMC article.
-
Candida auris and other phylogenetically related species - a mini-review of the literature.Germs. 2021 Sep 29;11(3):441-448. doi: 10.18683/germs.2021.1281. eCollection 2021 Sep. Germs. 2021. PMID: 34722366 Free PMC article. Review.
-
Rare fungal infectious agents: a lurking enemy.F1000Res. 2017 Oct 31;6:1917. doi: 10.12688/f1000research.11124.1. eCollection 2017. F1000Res. 2017. PMID: 29152230 Free PMC article. Review.
-
ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes.Front Microbiol. 2019 Jul 16;10:1445. doi: 10.3389/fmicb.2019.01445. eCollection 2019. Front Microbiol. 2019. PMID: 31379756 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

