Modeling the interplay between the HIF-1 and p53 pathways in hypoxia
- PMID: 26346319
- PMCID: PMC4561886
- DOI: 10.1038/srep13834
Modeling the interplay between the HIF-1 and p53 pathways in hypoxia
Abstract
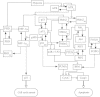
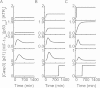
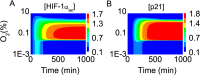
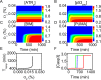
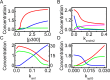
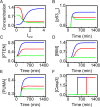
Both the hypoxia-inducible factor-1 (HIF-1) and tumor suppressor p53 are involved in the cellular response to hypoxia. How the two transcription factors interact to determine cell fates is less well understood. Here, we developed a network model to characterize crosstalk between the HIF-1 and p53 pathways, taking into account that HIF-1α and p53 are targeted for proteasomal degradation by Mdm2 and compete for binding to limiting co-activator p300. We reported the network dynamics under various hypoxic conditions and revealed how the stabilization and transcriptional activities of p53 and HIF-1α are modulated to determine the cell fate. We showed that both the transrepression and transactivation activities of p53 promote apoptosis induction. This work provides new insight into the mechanism for the cellular response to hypoxia.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

