The characteristics of Ishikawa endometrial cancer cells are modified by substrate topography with cell-like features and the polymer surface
- PMID: 26346435
- PMCID: PMC4531047
- DOI: 10.2147/IJN.S86336
The characteristics of Ishikawa endometrial cancer cells are modified by substrate topography with cell-like features and the polymer surface
Abstract
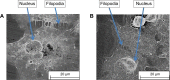
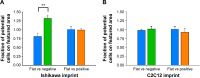
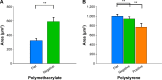
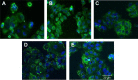
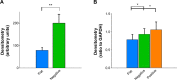
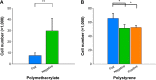
Conventional in vitro culture studies on flat surfaces do not reproduce tissue environments, which have inherent topographical mechanical signals. To understand the impact of these mechanical signals better, we use a cell imprinting technique to replicate cell features onto hard polymer culture surfaces as an alternative platform for investigating biomechanical effects on cells; the high-resolution replication of cells offers the micro- and nanotopography experienced in typical cell-cell interactions. We call this platform a Bioimprint. Cells of an endometrial adenocarcinoma cell line, Ishikawa, were cultured on a bioimprinted substrate, in which Ishikawa cells were replicated on polymethacrylate (pMA) and polystyrene (pST), and compared to cells cultured on flat surfaces. Characteristics of cells, incorporating morphology and cell responses, including expression of adhesion-associated molecules and cell proliferation, were studied. In this project, we fabricated two different topographies for the cells to grow on: a negative imprint that creates cell-shaped hollows and a positive imprint that recreates the raised surface topography of a cell layer. We used two different substrate materials, pMA and pST. We observed that cells on imprinted substrates of both polymers, compared to cells on flat surfaces, exhibited higher expression of β1-integrin, focal adhesion kinase, and cytokeratin-18. Compared to cells on flat surfaces, cells were larger on imprinted pMA and more in number, whereas on pST-imprinted surfaces, cells were smaller and fewer than those on a flat pST surface. This method, which provided substrates in vitro with cell-like features, enabled the study of effects of topographies that are similar to those experienced by cells in vivo. The observations establish that such a physical environment has an effect on cancer cell behavior independent of the characteristics of the substrate. The results support the concept that the physical topography of a cell's environment may modulate crucial oncological signaling pathways; this suggests the possibility of cancer therapies that target pathways associated with the response to mechanical stimuli.
Keywords: cell culture platforms; cell response; drug targets; mechanical forces; physical microenvironment; surface characteristics.
Figures


Similar articles
-
Cell-like features imprinted in the physical nano- and micro-topography of the environment modify the responses to anti-cancer drugs of endometrial cancer cells.Biofabrication. 2017 Feb 14;9(1):015017. doi: 10.1088/1758-5090/aa5c9a. Biofabrication. 2017. PMID: 28140336
-
Bioimprinted polymer platforms for cell culture using soft lithography.J Nanobiotechnology. 2014 Dec 30;12:60. doi: 10.1186/s12951-014-0060-6. J Nanobiotechnology. 2014. PMID: 25547467 Free PMC article.
-
The regulation of integrin-mediated osteoblast focal adhesion and focal adhesion kinase expression by nanoscale topography.Biomaterials. 2007 Apr;28(10):1787-97. doi: 10.1016/j.biomaterials.2006.12.020. Epub 2006 Dec 21. Biomaterials. 2007. PMID: 17218005
-
Controlling the morphology and outgrowth of nerve and neuroglial cells: The effect of surface topography.Acta Biomater. 2017 Mar 15;51:21-52. doi: 10.1016/j.actbio.2017.01.023. Epub 2017 Jan 7. Acta Biomater. 2017. PMID: 28069509 Review.
-
Nano-structured and functionalized surfaces for cytocompatibility improvement and bactericidal action.Biotechnol Adv. 2015 Nov 1;33(6 Pt 2):1120-9. doi: 10.1016/j.biotechadv.2015.01.001. Epub 2015 Jan 14. Biotechnol Adv. 2015. PMID: 25596482 Review.
Cited by
-
Tumour Initiation: a Discussion on Evidence for a "Load-Trigger" Mechanism.Cell Biochem Biophys. 2019 Dec;77(4):293-308. doi: 10.1007/s12013-019-00888-z. Epub 2019 Oct 9. Cell Biochem Biophys. 2019. PMID: 31598831 Free PMC article. Review.
-
Deciphering Biophysical Modulation in Ovarian Cancer Cells.Cell Biochem Biophys. 2021 Jun;79(2):375-386. doi: 10.1007/s12013-020-00964-9. Epub 2021 Jan 12. Cell Biochem Biophys. 2021. PMID: 33433760
-
Precision Medicine Gains Momentum: Novel 3D Models and Stem Cell-Based Approaches in Head and Neck Cancer.Front Cell Dev Biol. 2021 Jul 8;9:666515. doi: 10.3389/fcell.2021.666515. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34307351 Free PMC article. Review.
-
Regulation of cell fate by cell imprinting approach in vitro.Bioimpacts. 2024;14(3):29945. doi: 10.34172/bi.2023.29945. Epub 2023 Nov 28. Bioimpacts. 2024. PMID: 38938752 Free PMC article. Review.
-
Conductive Bioimprint Using Soft Lithography Technique Based on PEDOT:PSS for Biosensing.Bioengineering (Basel). 2021 Dec 9;8(12):204. doi: 10.3390/bioengineering8120204. Bioengineering (Basel). 2021. PMID: 34940357 Free PMC article.
References
-
- Chao EY, Inoue N. Biophysical stimulation of bone fracture repair, regeneration and remodelling. Eur Cell Mater. 2003;6:72–84. - PubMed
-
- Becker JL, Souza GR. Using space-based investigations to inform cancer research on Earth. Nat Rev Cancer. 2013;13:315–327. - PubMed
-
- Guo F, Li Y, Liu Y, et al. Identification of genes associated with tumor development in CaSki cells in the cosmic space. Mol Biol Rep. 2012;39:6923–6931. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

