Free fatty acid receptors: structural models and elucidation of ligand binding interactions
- PMID: 26346819
- PMCID: PMC4561419
- DOI: 10.1186/s12900-015-0044-2
Free fatty acid receptors: structural models and elucidation of ligand binding interactions
Abstract
Background: The free fatty acid receptors (FFAs), including FFA1 (orphan name: GPR40), FFA2 (GPR43) and FFA3 (GPR41) are G protein-coupled receptors (GPCRs) involved in energy and metabolic homeostasis. Understanding the structural basis of ligand binding at FFAs is an essential step toward designing potent and selective small molecule modulators.
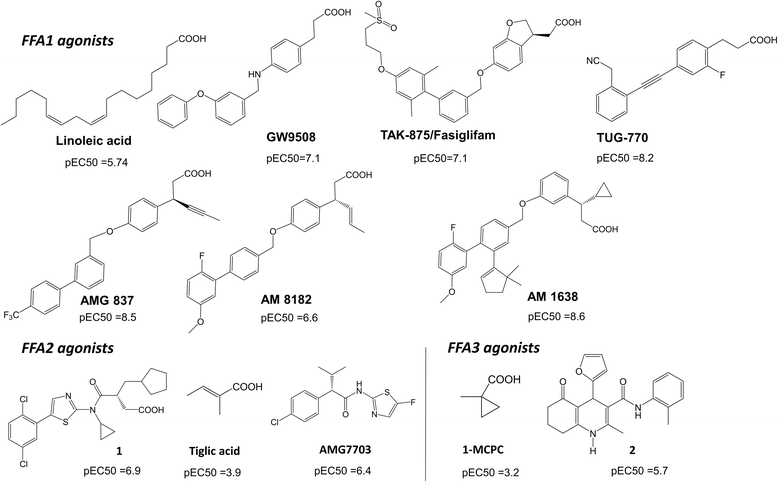
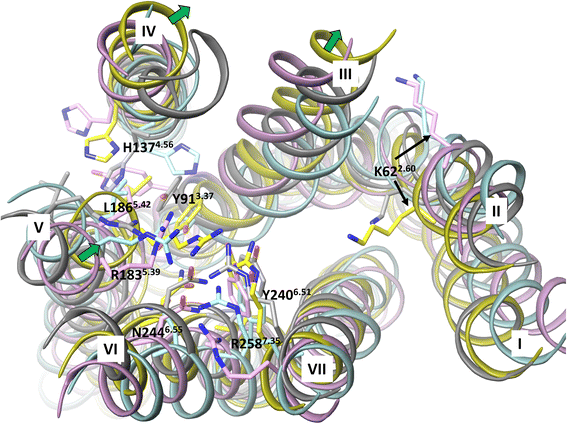
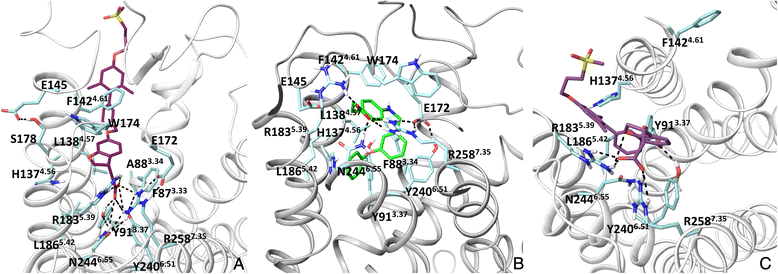
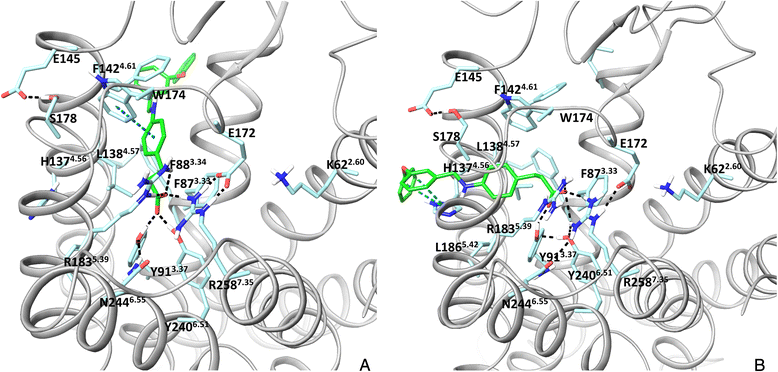
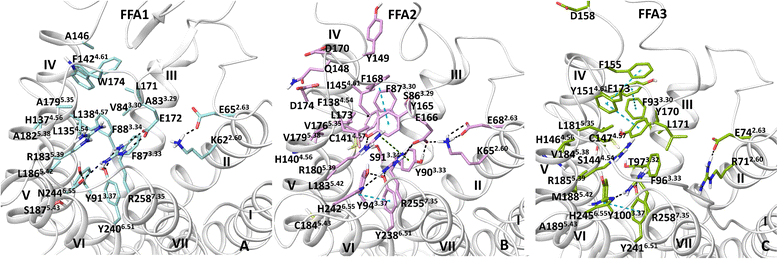
Results: We analyse earlier homology models of FFAs in light of the newly published FFA1 crystal structure co-crystallized with TAK-875, an ago-allosteric ligand, focusing on the architecture of the extracellular binding cavity and agonist-receptor interactions. The previous low-resolution homology models of FFAs were helpful in highlighting the location of the ligand binding site and the key residues for ligand anchoring. However, homology models were not accurate in establishing the nature of all ligand-receptor contacts and the precise ligand-binding mode. From analysis of structural models and mutagenesis, it appears that the position of helices 3, 4 and 5 is crucial in ligand docking. The FFA1-based homology models of FFA2 and FFA3 were constructed and used to compare the FFA subtypes. From docking studies we propose an alternative binding mode for orthosteric agonists at FFA1 and FFA2, involving the interhelical space between helices 4 and 5. This binding mode can explain mutagenesis results for residues at positions 4.56 and 5.42. The novel FFAs structural models highlight higher aromaticity of the FFA2 binding cavity and higher hydrophilicity of the FFA3 binding cavity. The role of the residues at the second extracellular loop used in mutagenesis is reanalysed. The third positively-charged residue in the binding cavity of FFAs, located in helix 2, is identified and predicted to coordinate allosteric modulators.
Conclusions: The novel structural models of FFAs provide information on specific modes of ligand binding at FFA subtypes and new suggestions for mutagenesis and ligand modification, guiding the development of novel orthosteric and allosteric chemical probes to validate the importance of FFAs in metabolic and inflammatory conditions. Using our FFA homology modelling experience, a strategy to model a GPCR, which is phylogenetically distant from GPCRs with the available crystal structures, is discussed.
Figures


Similar articles
-
Application of GPCR Structures for Modelling of Free Fatty Acid Receptors.Handb Exp Pharmacol. 2017;236:57-77. doi: 10.1007/164_2016_52. Handb Exp Pharmacol. 2017. PMID: 27757764
-
Selective orthosteric free fatty acid receptor 2 (FFA2) agonists: identification of the structural and chemical requirements for selective activation of FFA2 versus FFA3.J Biol Chem. 2011 Mar 25;286(12):10628-40. doi: 10.1074/jbc.M110.210872. Epub 2011 Jan 10. J Biol Chem. 2011. PMID: 21220428 Free PMC article.
-
Agonism and allosterism: the pharmacology of the free fatty acid receptors FFA2 and FFA3.Br J Pharmacol. 2009 Sep;158(1):146-53. doi: 10.1111/j.1476-5381.2009.00421.x. Br J Pharmacol. 2009. PMID: 19719777 Free PMC article. Review.
-
Investigation of the Binding Interaction of Fatty Acids with Human G Protein-Coupled Receptor 40 Using a Site-Specific Fluorescence Probe by Flow Cytometry.Biochemistry. 2016 Apr 5;55(13):1989-96. doi: 10.1021/acs.biochem.6b00079. Epub 2016 Mar 17. Biochemistry. 2016. PMID: 26974599
-
Role of free fatty acid receptors in the regulation of energy metabolism.Biochim Biophys Acta. 2014 Sep;1841(9):1292-300. doi: 10.1016/j.bbalip.2014.06.002. Epub 2014 Jun 10. Biochim Biophys Acta. 2014. PMID: 24923869 Review.
Cited by
-
Olive Oil Lipophenols Induce Insulin Secretion in 832/13 β-Cell Models.Pharmaceutics. 2021 Jul 16;13(7):1085. doi: 10.3390/pharmaceutics13071085. Pharmaceutics. 2021. PMID: 34371780 Free PMC article.
-
A single extracellular amino acid in Free Fatty Acid Receptor 2 defines antagonist species selectivity and G protein selection bias.Sci Rep. 2017 Oct 23;7(1):13741. doi: 10.1038/s41598-017-14096-3. Sci Rep. 2017. PMID: 29061999 Free PMC article.
-
Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.Nat Struct Mol Biol. 2017 Jul;24(7):570-577. doi: 10.1038/nsmb.3417. Epub 2017 Jun 5. Nat Struct Mol Biol. 2017. PMID: 28581512
-
Analysis of microbiota-host communication mediated by butyrate in Atlantic salmon.Comput Struct Biotechnol J. 2023 Mar 31;21:2558-2578. doi: 10.1016/j.csbj.2023.03.050. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37122632 Free PMC article.
-
Short-Chain Fatty Acid Receptors and Cardiovascular Function.Int J Mol Sci. 2022 Mar 18;23(6):3303. doi: 10.3390/ijms23063303. Int J Mol Sci. 2022. PMID: 35328722 Free PMC article. Review.
References
-
- Briscoe CP, Peat AJ, McKeown SC, Corbett DF, Goetz AS, Littleton TR, McCoy DC, Kenakin TP, Andrews JL, Ammala C, Fornwald JA, Ignar DM, Jenkinson S. Pharmacological regulation of insulin secretion in MIN6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small molecules. Br J Pharmacol. 2006;148(5):619–628. doi: 10.1038/sj.bjp.0706770. - DOI - PMC - PubMed
-
- Itoh Y, Hinuma S. GPR40, a free fatty acid receptor on pancreatic beta cells, regulates insulin secretion. Hepatol Res. 2005;33(2):171–173. - PubMed
-
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278(13):11312–11319. doi: 10.1074/jbc.M211609200. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

