Insights into the bacterial community and its temporal succession during the fermentation of wine grapes
- PMID: 26347718
- PMCID: PMC4539513
- DOI: 10.3389/fmicb.2015.00809
Insights into the bacterial community and its temporal succession during the fermentation of wine grapes
Abstract
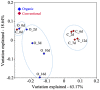
Grapes harbor complex microbial communities. It is well known that yeasts, typically Saccharomyces cerevisiae, and bacteria, commonly the lactic acid fermenting Oenococcus oeni, work sequentially during primary and secondary wine fermentation. In addition to these main players, several microbes, often with undesirable effects on wine quality, have been found in grapes and during wine fermentation. However, still little is known about the dynamics of the microbial community during the fermentation process. In previous studies culture dependent methods were applied to detect and identify microbial organisms associated with grapes and grape products, which resulted in a picture that neglected the non-culturable fraction of the microbes. To obtain a more complete picture of how microbial communities change during grape fermentation and how different fermentation techniques might affect the microbial community composition, we employed next-generation sequencing (NGS)-a culture-independent method. A better understanding of the microbial dynamics and their effect on the final product is of great importance to help winemakers produce wine styles of consistent and high quality. In this study, we focused on the bacterial community dynamics during wine vinification by amplifying and sequencing the hypervariable V1-V3 region of the 16S rRNA gene-a phylogenetic marker gene that is ubiquitous within prokaryotes. Bacterial communities and their temporal succession was observed for communities associated with organically and conventionally produced wines. In addition, we analyzed the chemical characteristics of the grape musts during the organic and conventional fermentation process. These analyses revealed distinct bacterial population with specific temporal changes as well as different chemical profiles for the organically and conventionally produced wines. In summary these results suggest a possible correlation between the temporal succession of the bacterial population and the chemical wine profiles.
Keywords: 16S rRNA gene profile; next-generation sequencing; organic grape products; temporal succession; wine bacteria; wine fermentation.
Figures


Similar articles
-
The microbial ecology of wine grape berries.Int J Food Microbiol. 2012 Feb 15;153(3):243-59. doi: 10.1016/j.ijfoodmicro.2011.11.025. Epub 2011 Dec 2. Int J Food Microbiol. 2012. PMID: 22189021
-
Lactic acid bacteria in the quality improvement and depreciation of wine.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):317-31. Antonie Van Leeuwenhoek. 1999. PMID: 10532386 Review.
-
Microbiome dynamics during spontaneous fermentations of sound grapes in comparison with sour rot and Botrytis infected grapes.Int J Food Microbiol. 2018 Sep 20;281:36-46. doi: 10.1016/j.ijfoodmicro.2018.05.016. Epub 2018 May 18. Int J Food Microbiol. 2018. PMID: 29807290
-
Next-generation sequencing reveals significant bacterial diversity of botrytized wine.PLoS One. 2012;7(5):e36357. doi: 10.1371/journal.pone.0036357. Epub 2012 May 1. PLoS One. 2012. PMID: 22563494 Free PMC article.
-
The Whiff of Wine Yeast Innovation: Strategies for Enhancing Aroma Production by Yeast during Wine Fermentation.J Agric Food Chem. 2019 Dec 11;67(49):13496-13505. doi: 10.1021/acs.jafc.9b06191. Epub 2019 Nov 25. J Agric Food Chem. 2019. PMID: 31724402 Review.
Cited by
-
Exploring functional core bacteria in fermentation of a traditional Chinese food, Aspergillus-type douchi.PLoS One. 2019 Dec 30;14(12):e0226965. doi: 10.1371/journal.pone.0226965. eCollection 2019. PLoS One. 2019. PMID: 31887171 Free PMC article.
-
Metataxonomic Analysis of Grape Microbiota During Wine Fermentation Reveals the Distinction of Cyprus Regional terroirs.Front Microbiol. 2021 Sep 22;12:726483. doi: 10.3389/fmicb.2021.726483. eCollection 2021. Front Microbiol. 2021. PMID: 34630353 Free PMC article.
-
DNA sequencing, genomes and genetic markers of microbes on fruits and vegetables.Microb Biotechnol. 2021 Mar;14(2):323-362. doi: 10.1111/1751-7915.13560. Epub 2020 Mar 24. Microb Biotechnol. 2021. PMID: 32207561 Free PMC article. Review.
-
Geographical and Cultivar Features Differentiate Grape Microbiota in Northern Italy and Spain Vineyards.Front Microbiol. 2018 May 15;9:946. doi: 10.3389/fmicb.2018.00946. eCollection 2018. Front Microbiol. 2018. PMID: 29867854 Free PMC article.
-
The Grapevine and Wine Microbiome: Insights from High-Throughput Amplicon Sequencing.Front Microbiol. 2017 May 11;8:820. doi: 10.3389/fmicb.2017.00820. eCollection 2017. Front Microbiol. 2017. PMID: 28553266 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

